Crambe filiformis fatty acid elongase and coding gene thereof
A fatty acid elongase, encoding gene technology, applied in the fields of molecular biology and genetic engineering, can solve the problem of complex erucic acid content
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
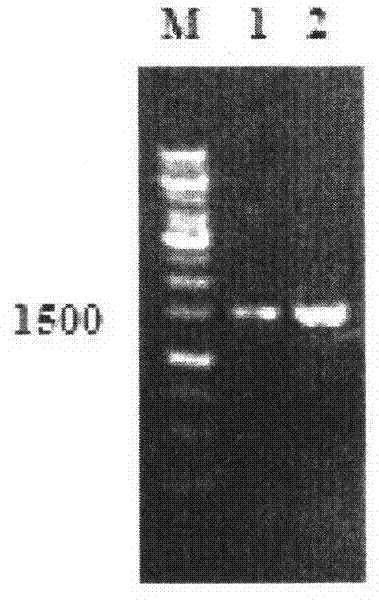
[0016] Embodiment 1, the cloning of gene CfFAE1
[0017] Clone CfFAE1, the specific steps are as follows:
[0018] 1. Acquisition of CfFAE1 gene
[0019] C. filiformis was cultivated under normal conditions. After the true leaves grew, the leaves were collected, DNA was extracted from the leaves, polymerase chain reaction was carried out, and finally the CfFAE1 gene was obtained.
[0020] 1. Extraction of C. filiformis genomic DNA
[0021] Take about 0.2 g of fresh young living plant leaves, grind them fully under liquid nitrogen freezing conditions, transfer them to a 1.5 mL centrifuge tube, add 500 μL of CTAB extract, and bathe in water at 65°C for 45 minutes, during which time they are inverted and mixed 3 times; add 500 μL of Chloroform-isoamyl alcohol with a volume ratio of 24:1, mixed evenly, 12000r·min -1 Centrifuge for 15 min; take the supernatant, add 2 times the volume of absolute ethanol, and put it in the refrigerator at -20°C overnight; 4000r min -1 Centrifuge...
Embodiment 2
[0031] Example 2, Functional Verification of CfFAE1 Gene
[0032] 1. High expression of CfFAE1 gene in yeast
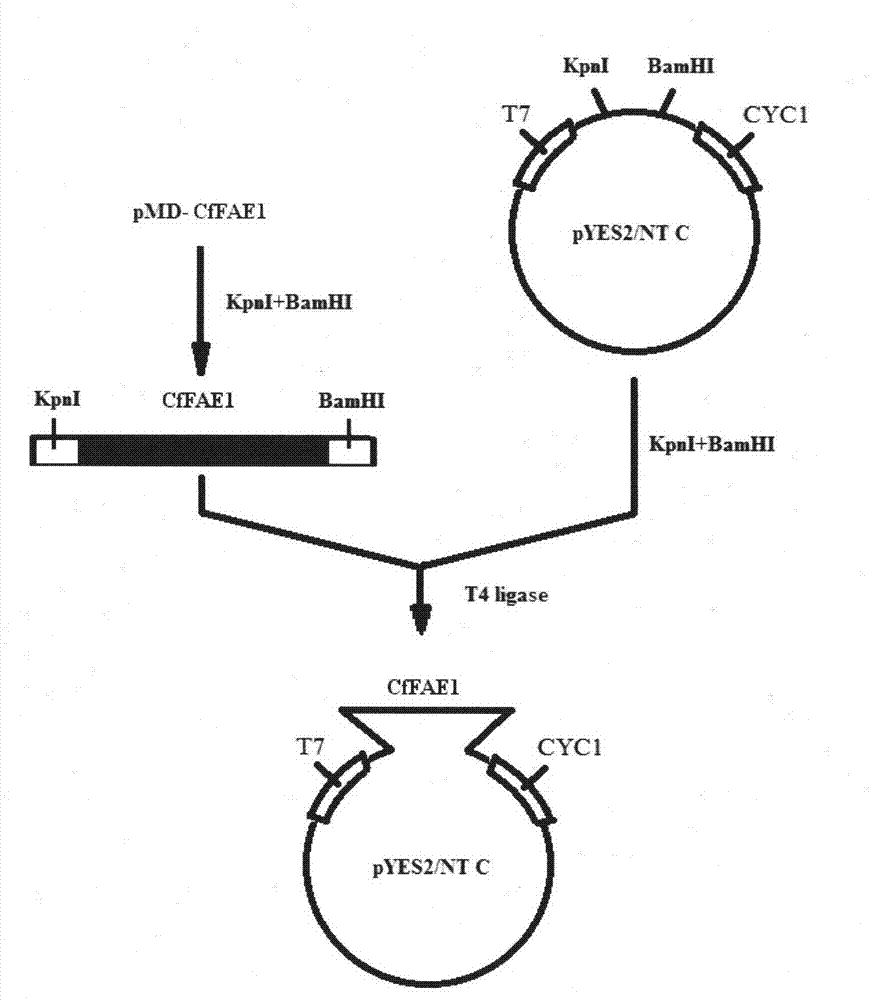
[0033] In order to demonstrate the coding function of the CfFAE1 gene, the present invention clones the CfFAE1 gene between the BamHI and KpnI sites of pYES2 / NT C from Invitrogen, and transforms the yeast strain InvSc1 to obtain high-level expression.
[0034] The present invention utilizes the multiple cloning sites of the pYES2 / NT C high-efficiency expression vector of Invitrogen Company to realize the high-efficiency expression of CfFAE1. The enzyme cleavage site used in this experiment makes the 5' end of the product expressed by the pYES2 / NT C expression vector add 6 consecutive His fusion proteins, and these 6 consecutive His constitute the specific binding site of i-NTA gel. Therefore, the expression product can be purified by affinity chromatography.
[0035] The pMD-CfFAE1 recombinant vector carrying the CfFAE1 gene and the pYES2 / NT C expression vector were...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com