Sequence-selective recognition of nucleic acids using nanoparticle probes
A technology of nanoparticle and target nucleic acid, applied in nanotechnology for sensing, nanotechnology, nanotechnology, etc., which can solve the problems of difficult preparation and low solubility of PNA/nanoparticle conjugates
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0119] Example 1: Materials
[0120] Chemical reagent: tetrachloroauric(III) acid trihydrate (HAuCl 4 ·3H 2 O), trisodium citrate, TritonX-100, sodium dodecyl sulfate (SDS), ZonylFSN-100 (F(CF 2 CF 2 ) 3-8 CH 2 CH 2 O(CH 2 CH 2 O) 0-15 H), tris(2-carboxyethyl)phosphine (TCEP) and dithiothreitol (DTT) were purchased from Sigma-Aldrich (St Louis, MO). 40-nm silver nanoparticles were purchased from TedPella (Redding, CA). All other reagents certified as analytically pure were used as received. Morpholino oligomers were purchased from GeneTools LLC (Philomath, OR). oligonucleotides from 1 st Acquired from Base Pte Ltd (Singapore).
[0121] Instrument: with NanoDrop TM A 1000 spectrophotometer (Thermo Scientific) was used for quantification of morpholino oligonucleotides and ssDNA samples. Agilent G1103AUV-Vis spectrophotometer was used to collect the absorption spectrum of gold nanoparticle colloid. This spectrophotometer was also used for melting analysis. Absorb...
Embodiment 2
[0122] Example 2: Synthesis of gold nanoparticles
[0123] By reducing HAuCl with citrate as described by Grabar, K.C. et al., Anal.Chem., 1995, 67, 735-743 4 to prepare gold nanoparticles with average diameters of ~40 nm and ~13 nm, respectively. All glassware used to prepare gold nanoparticles was sequentially filled with freshly prepared aqua regia (HNO 3 :HCl=1:3), rinse thoroughly with ultrapure water, and then dry in an oven at 100°C for 2-3 hours. 60 mL of 0.01% (w / v) HAuCl 4 Boil in a round bottom flask with reflux condenser and stir vigorously. For gold nanoparticles with an average diameter of ~40 nm, add 0.6 mL of 1.0% (w / v) sodium citrate to HAuCl 4 solution. For gold nanoparticles with an average diameter of ~13 nm, add 4.5 mL of 1.0 wt% sodium citrate to HAuCl 4 in solution. The reaction mixture was maintained at boiling point and stirring was continued for about 15 minutes. The suspension was stored at 4°C until subsequent use. Assuming spherical partic...
Embodiment 3
[0124] Example 3: Preparation of metal nanoparticles modified with mercapto-morpholino oligonucleotides
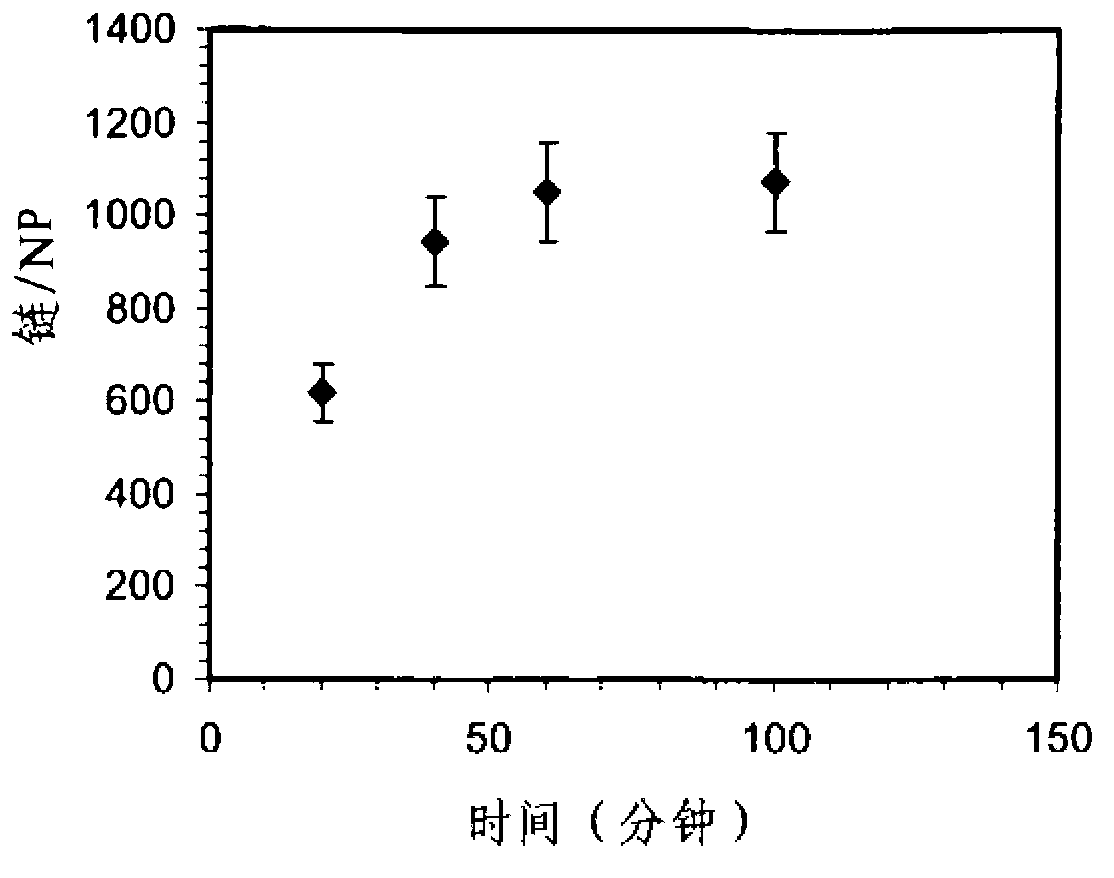
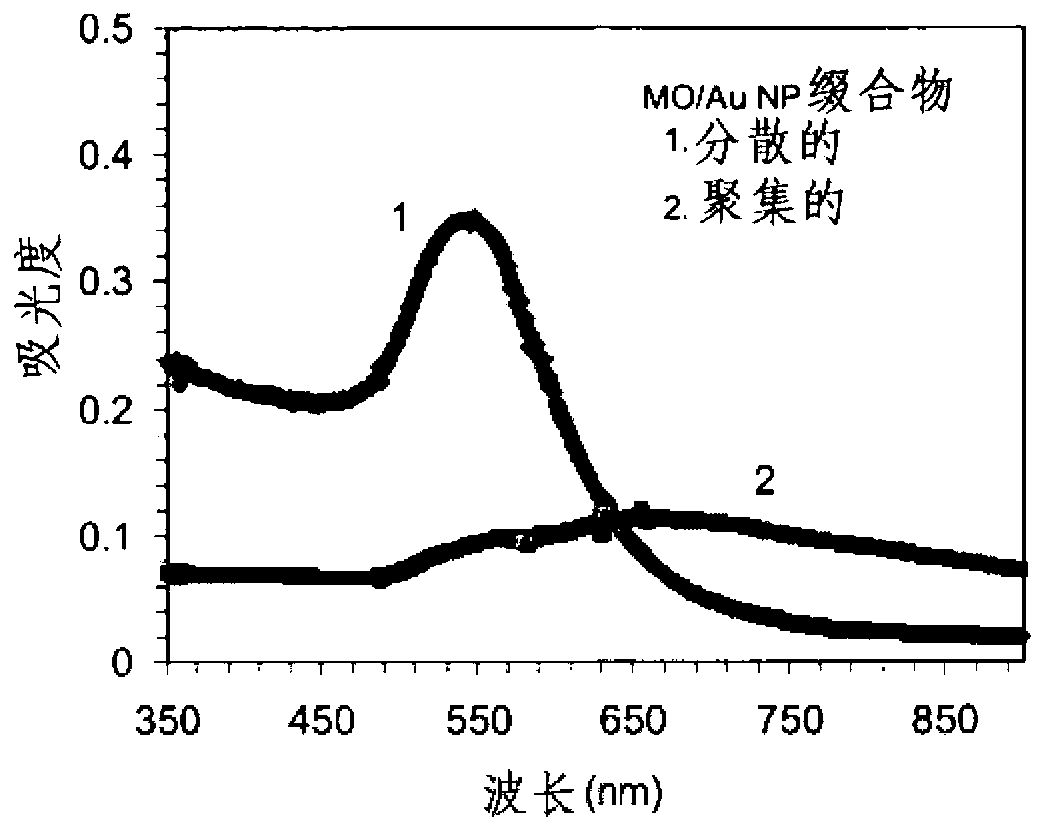
[0125]By activating the sulfhydryl group, the 3'-disulfamide-modified MO was treated with 0.1 MDTT in 0.2M phosphate buffered saline (PBS, pH 8.0) for 1 hour; the thiolated MO was then purified using a NAP-5 column (GE Healthcare). The purified thiolated MO samples were stored at 4°C until subsequent use. To avoid interchain disulfide formation of the thiolated MO chains, the purified MO was dissolved in 5 mM tris(2-carboxyethyl)phosphine (TCEP) (pH 7.5) before mixing with gold or silver nanoparticles. Incubate for 10 minutes. Unless otherwise stated, the mixture solution containing ˜2 μM thiolated MO, ˜2 nM or 4 nM FSN-capped nanoparticles, ˜0.1 wt % SDS, and 10 mM phosphate buffer (pH 7.5) was incubated at room temperature for 1 h or 2 h. Hour. Then, excess ssDNA was removed by centrifugation at 7.0 Krpm for 10 minutes. Unreacted MO was removed by centrifugation at 1...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Surface charge | aaaaa | aaaaa |
| Tm value | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com