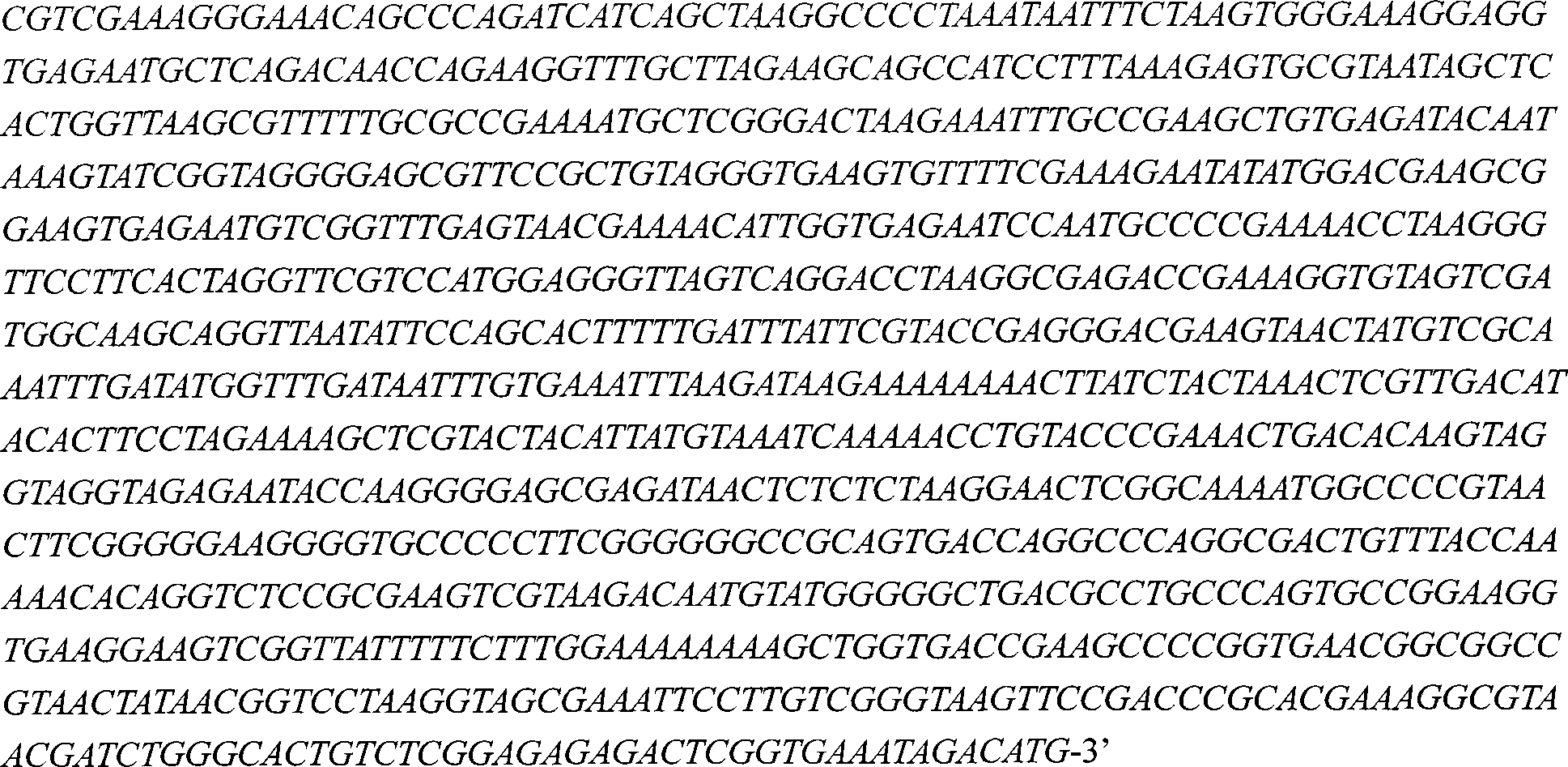Method for constructing Platymonas subcordiformis chloroplast expression system
A chloroplast expression and subcardioid technology, applied in the field of biological genetic engineering, can solve the problem of limited genetic transformation of subcardioid flat algae, and achieve the effects of stable expression, high expression and high expression efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0032] Example: Achieve stable expression of herbicide resistance gene in Platymonas subcordiformis chloroplast
[0033] 1. Amplification and cloning of two chloroplast homologous recombination fragments of Platymonas subcordiformis
[0034] Design and synthesize the following two pairs of primers
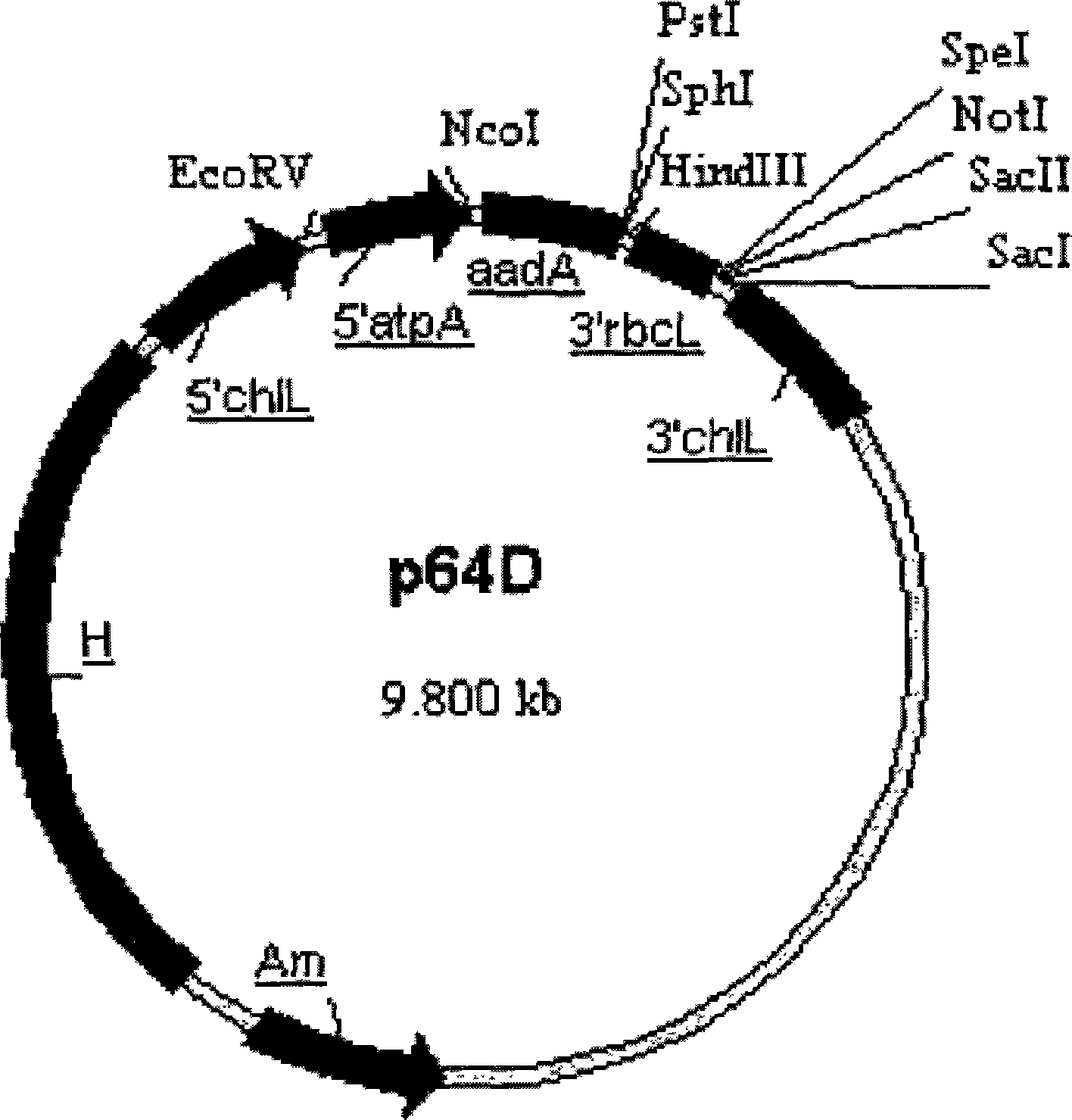
[0035] P1: 5’-CGG GGTACC TCCTACGGGAGGCAGCAGT-3' (underline is Kpn I restriction site)
[0036] P2: 5’-GC GTCGAC TTGAGGCAAATGGGCTATGC-3' (underline is Sal I restriction site)
[0037] P3: 5’-TT GCGGCCGC ACAACGGAGTTTCGGAATA-3' (underline is Not I restriction site)
[0038] P4: 5’-C GAGCTC CGTTACTCAAACCGACATTC-3' (Sac I restriction site is underlined)
[0039] The amplification product of primers P1 and P2 is SEQ ID NO: 1, which is the fragment 16S-trnI; the amplification product of primers P3 and P4 is SEQ ID NO: 2, which is the fragment trnA-23S (see figure 1 ).
[0040] Using the total DNA of Platymonas subcordiformis as a template, PCR amplification was carried out with primers P1 and P2. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com