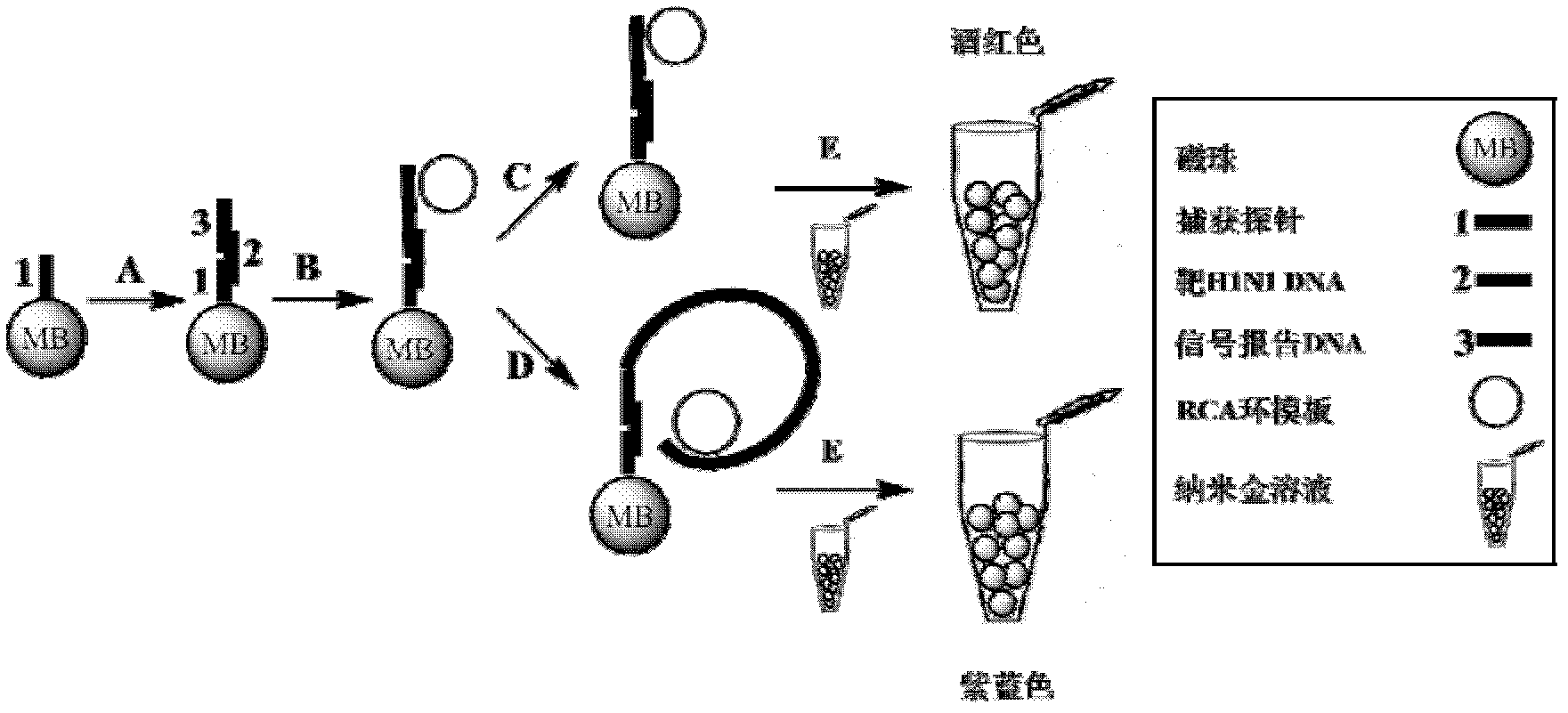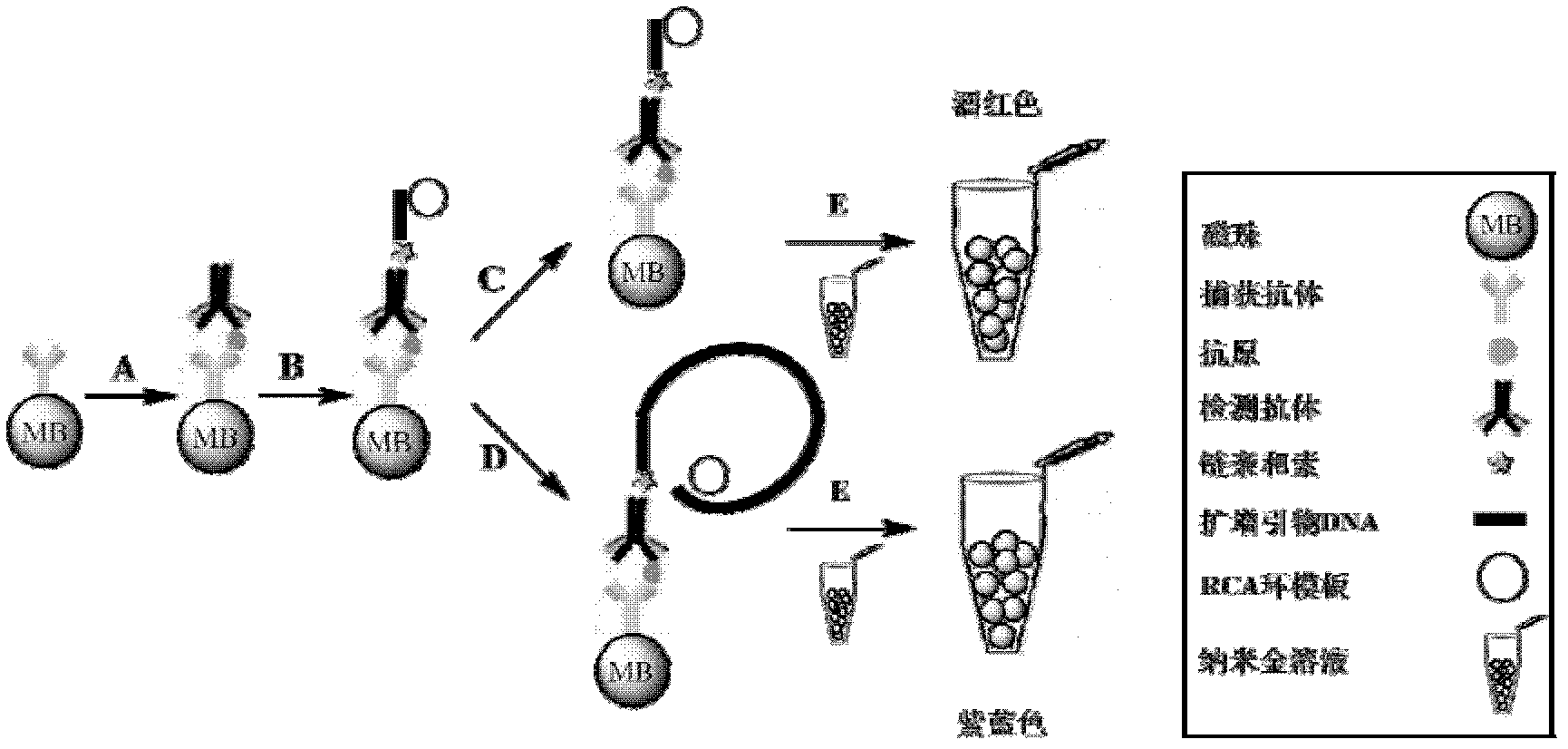Rolling circle amplification-based colorimetric assay method for target nucleic acids or proteins
A technology of rolling circle amplification and target nucleic acid, applied in biochemical equipment and methods, biological testing, material inspection products, etc., can solve problems such as inability to adapt to rapid detection and complexity, and achieve intuitive and clear detection results, simple operation, and specificity good sex effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
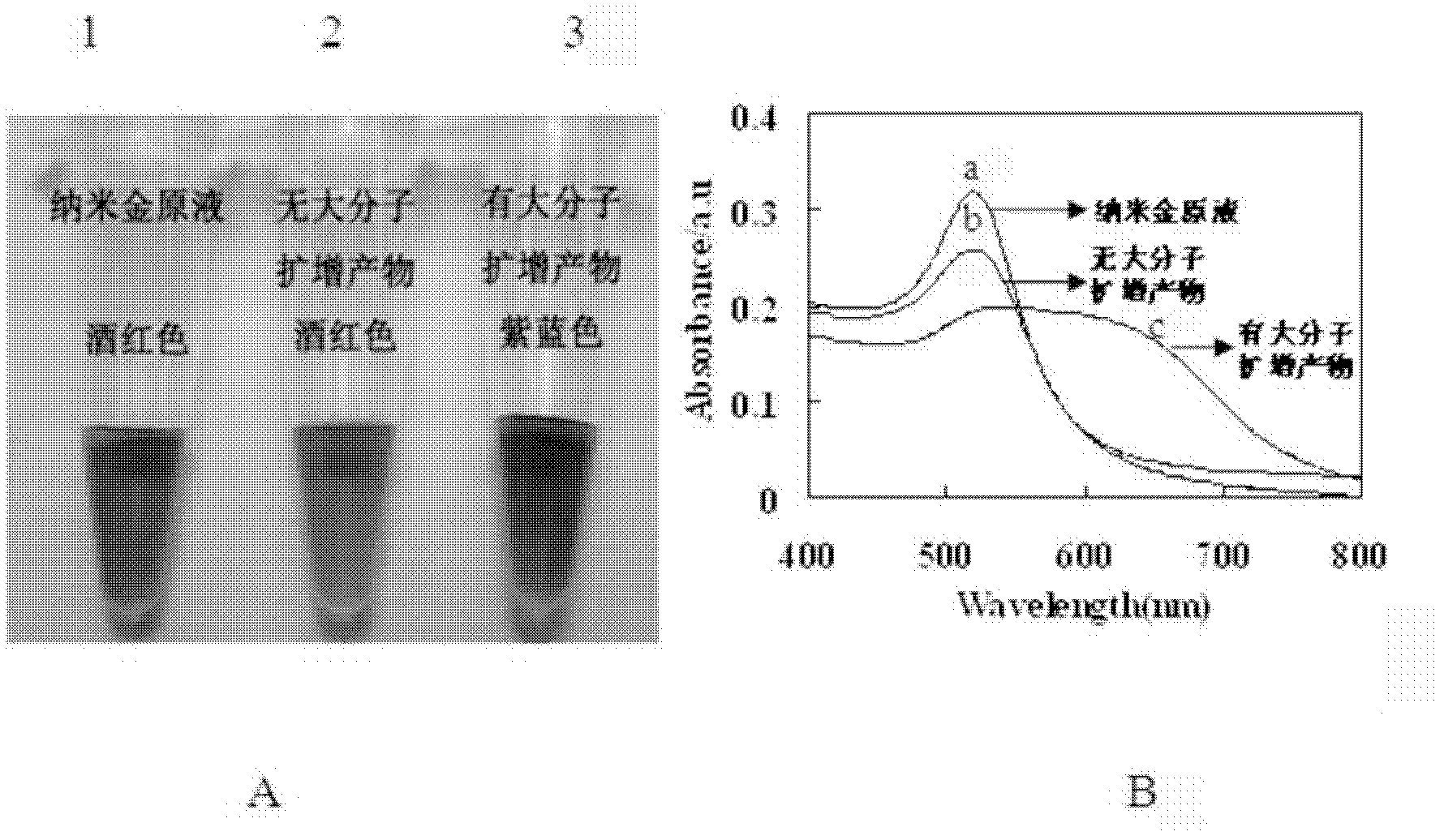
[0039] A colorimetric detection method for target nucleic acid based on rolling circle amplification
[0040] This example describes the establishment of a method for colorimetric detection of target nucleic acids based on rolling circle amplification, taking type A H1N1 nucleic acids as an example
[0041] (1) Design and synthesis of detection probes
[0042] The present invention uses the influenza virus H1N1 (Influenza A virus H1N1envelope_2) sequence as the target gene template, and the DNA probe sequence is as follows (the synthesis of the probe is completed by Dalian TaKaRa Biotechnology Co., Ltd.).
[0043] Table 1. H1N1 DNA target sequences and probes
[0044]
[0045] (2) Labeling of capture probes on magnetic beads
[0046] Take 50 μL of unlabeled amino magnetic beads, add 50 μL of MES (0.1mol / L, pH 4.6), wash twice, magnetically separate, and discard the supernatant. Add 40 μl of MES and 10 μl of carboxy-modified capture probe, and mix thoroughly. Prepare 100...
Embodiment 2
[0055] Colorimetric detection of target protein based on rolling circle amplification
[0056] This example describes the establishment of a method for colorimetric detection of target proteins based on rolling circle amplification, taking the heart disease marker troponin as an example
[0057] 1. Design and synthesis of signal reporter primer probe
[0058]
[0059] 2 Labeling of capture antibodies on magnetic beads
[0060] Wash 50ul magnetic beads with an equal amount of MES (25Mm, PH6) twice, remove the supernatant after magnetic separation, add 25ul EDC and NHS (freshly prepared, the concentration is 50mg / ml) in turn, mix and incubate for 30 minutes, magnetic After separation, remove the supernatant and wash twice with MES; dissolve the activated magnetic beads in 45ul MES, mix well with 2.5ug antibody, and incubate for 30 minutes (slow speed to prevent magnetic beads from sedimenting); then add 50ul Tris (PH7 , 50mM) for 30 minutes to seal the unreacted groups on t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com