Method for identifying pennisetum forage grass by utilizing ISSR (Inter Simple Sequence Repeat) molecular marker technology
A molecular marker and technical identification technology, which is applied in the field of forage identification using molecular marker technology to achieve the effect of ensuring vigorous promotion, reducing workload and ensuring authenticity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Screening of ISSR molecular markers of Pennisetum
[0037] (1) Based on the collected materials of 2 main species of Pennisetum and their hybrids, they are: Hybrid Pennisetum No. 1, Hybrid Pennisetum No. 2, and Hybrid Pennisetum No. 3 , Elephant Grass, Hybrid Pennisetum, Purple Pennisetum, Broadleaf Pennisetum;
[0038] (2) Seedling cultivation: select 100 uniform seeds of each variety, and cultivate them for 20 days under greenhouse conditions to form normal seedlings as test materials;
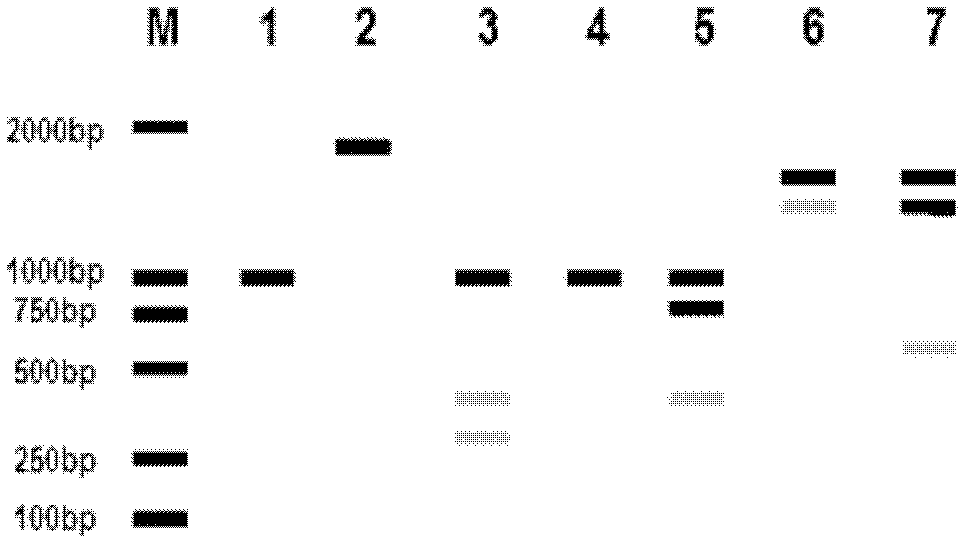
[0039] (3) A new type of plant genome DNA extraction kit (Tiangen Reagent Company) was used to extract the total DNA of a single seedling, and three single plants were randomly selected for each material, and 0.8% agarose gel electrophoresis was used for detection and DNA was determined with a nucleic acid protein analyzer. The concentration of the sample is shown in Table 1, and the DNA pool of each material was prepared so that the final concentration of DNA was 10 ng / μL;...
Embodiment 2
[0055] Example 2 Identification method 1
[0056] (1) Extract Pennisetum DNA and construct a DNA pool;
[0057] Select the seven Pennisetum materials that need to be identified: Hybrid Pennisetum No. 1, Hybrid Pennisetum No. 3, Hybrid Pennisetum No. 3, Elephant Grass, Hybrid Pennisetum, Ziguang Pennisetum, and Broad-leaved Pennisetum For one or more of them, select 100 uniform seeds from each material to be identified, and cultivate them under greenhouse conditions for 20 days to form normal seedlings. Take the leaves of a single seedling and use Tiangen’s new plant genome DNA extraction reagent Genomic DNA of Pennisetum genus was extracted from each box, and three individual plants were extracted from each material, detected by 0.8% agarose gel electrophoresis, and the DNA sample concentration was determined by a nucleic acid and protein analyzer. Three individual plants of each material were used The DNA was mixed and diluted in equal amounts to prepare DNA pools for the DN...
Embodiment 3
[0064] Example 3 Identification method 2
[0065] (1) Extract Pennisetum DNA and construct a DNA pool;
[0066] Select the seven Pennisetum materials that need to be identified: Hybrid Pennisetum No. 1, Hybrid Pennisetum No. 3, Hybrid Pennisetum No. 3, Elephant Grass, Hybrid Pennisetum, Ziguang Pennisetum, and Broad-leaved Pennisetum For one or more of them, select 100 uniform seeds from each material to be identified, and cultivate them under greenhouse conditions for 20 days to form normal seedlings. Take the leaves of a single seedling and use Tiangen’s new plant genome DNA extraction reagent Genomic DNA of Pennisetum genus was extracted from each box, and three individual plants were extracted from each material, detected by 0.8% agarose gel electrophoresis, and the DNA sample concentration was determined by a nucleic acid and protein analyzer. Three individual plants of each material were used The DNA was mixed and diluted in equal amounts to prepare DNA pools for the DN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com