Preparation method for probes related to breast cancer molecular markers and application of same
A probe and gene probe technology, applied in the preparation of breast cancer molecular marker-related probes and their application fields, can solve the problems of decreased curative effect, shortened recurrence-free survival period, poor patient prognosis, etc., and improved the detection rate , Accurate breast cancer classification, simple operation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Embodiment 1: Preparation of HER2 gene probe
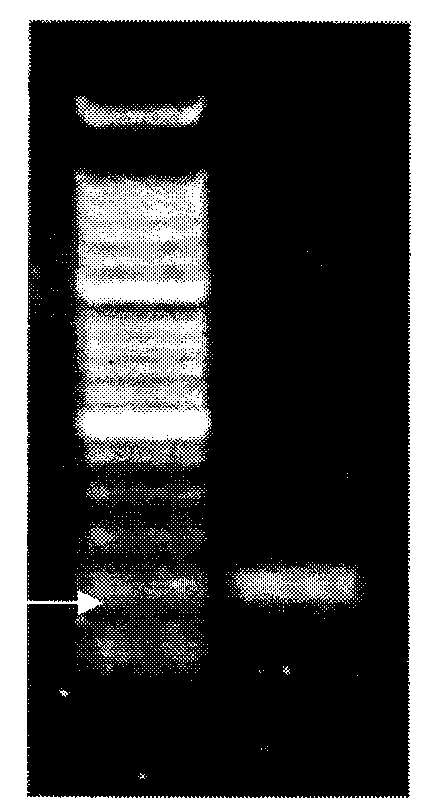
[0047] (1) Primer design and clone screening: HER2 gene is located in the 17q12 segment of human chromosome. NCBI Mapview database searches all clones containing HER2 gene, and screens out the clone containing this gene, numbered RP11-909L6.
[0048] (2) Cloning culture and identification: Purchase the clone RP11-909L6, take 10 μl of the cloning bacteria solution and add it to 5 ml of TB culture solution (chloramphenicol resistance), and shake the bacteria in a shaker at 37°C for 8 to 12 hours; Add all the solution to 500ml TB culture solution (chloramphenicol resistance), shake the bacteria in a shaker at 37°C for 8-12 hours; the bacteria solution uses upstream primers
[0049] 5'-AGGGGAGAATAAATAAAATCTGTGG-3' and downstream primers
[0050] 5'-CAGGAGTGAGACACTCTCCATG-3' was amplified by PCR, and the amplification conditions were: 95°C for 5 minutes; (94°C for 30 seconds, 56°C for 30 seconds, 72°C for 45 seconds) × 40 cycle...
Embodiment 2
[0070] Embodiment 2: the preparation of TOP2A gene probe
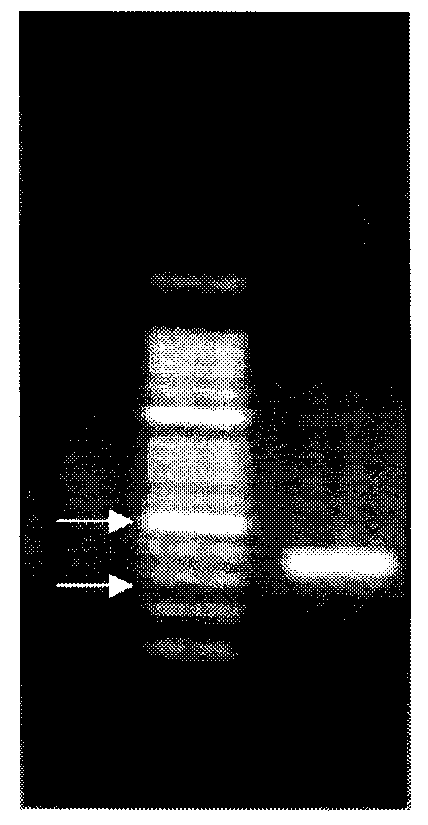
[0071] (1) Primer design and clone screening: The TOP2A gene is located on the 17q21 segment of human chromosome, and all clones containing the TOP2A gene were searched in the NCBI Mapview database, and the clone containing the gene was screened out, numbered RP11-1029L16.
[0072] (2) Cloning culture and identification: purchase the clone RP11-1029L16, take 10 μl of the cloning bacteria solution and add it to 5ml TB culture solution (chloramphenicol resistance), and shake the bacteria in a shaking table at 37°C for 8-12 hours; Add all the solution to 500ml TB culture solution (chloramphenicol resistance), shake the bacteria in a shaker at 37°C for 8-12 hours; use the upstream primer 5'-ATTCAAAGCTGGATCCCTTT-3' and the downstream primer 5'-AGCTGTGACAAATGCCTGTA for the bacterial solution -3' was subjected to PCR amplification, and the amplification conditions were: 95° C. for 5 minutes; (94° C. for 30 seconds, 56° C. for...
Embodiment 3
[0092] Embodiment 3: the preparation of AGTR1 gene probe
[0093] (1) Primer design and clone screening: AGTR1 gene is located in the 3q24 segment of human chromosome. NCBI Mapview database searched all clones containing AGTR1 gene, and screened out the clone containing this gene, numbered RP11-366P9.
[0094] (2) Cloning culture and identification: purchase the clone RP11-366P9, take 10 μl of the cloning bacteria solution and add it to 5ml TB culture solution (chloramphenicol resistance), and shake the bacteria in a shaker at 37°C for 8 to 12 hours; Add all the solution to 500ml TB culture solution (chloramphenicol resistance), shake the bacteria in a shaker at 37°C for 8-12 hours; use the upstream primer 5'-TTGCATTATTTTCAAAGCCCTT-3' and the downstream primer 5'-TCCTACCAGCTTAGGCCAGA for the bacterial solution -3' was subjected to PCR amplification, and the amplification conditions were: 95° C. for 5 minutes; (94° C. for 30 seconds, 56° C. for 30 seconds, 72° C. for 45 seconds...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com