Application of biphenyl dioxygenase in degrading polychlorinated biphenyl
A technology of biphenyl dioxygenase and hydroxybiphenyl dioxygenase, which is applied in the application field of biphenyl dioxygenase in the degradation of polychlorinated biphenyls, can solve the problems of low degradation efficiency and improve the degradation capacity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Construction of Pseudomonas fluorescens expression gene vector with optimized codon-biased dihydroxybiphenyl dioxygenase gene
[0022] (1) Test method:
[0023] The characteristics of the Pseudomonas fluorescens expression vector pFB1690 with pGM promoter and t1t2 terminator constructed by the Genetic Engineering Research Office of Biotechnology Research Institute of Shanghai Academy of Agricultural Sciences are as follows: the expression of the target gene is controlled by the pGM promoter and t1t2 terminator, Km resistance gene as a selection marker.
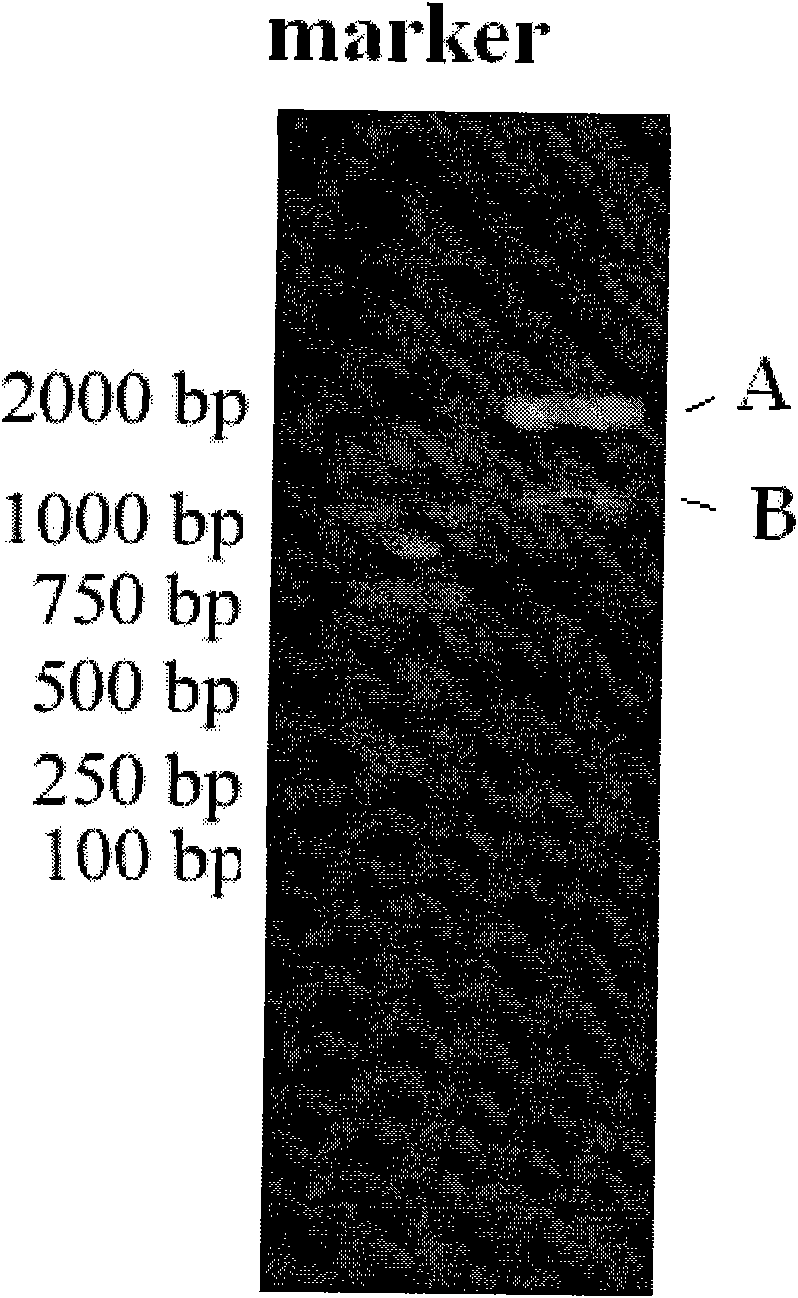
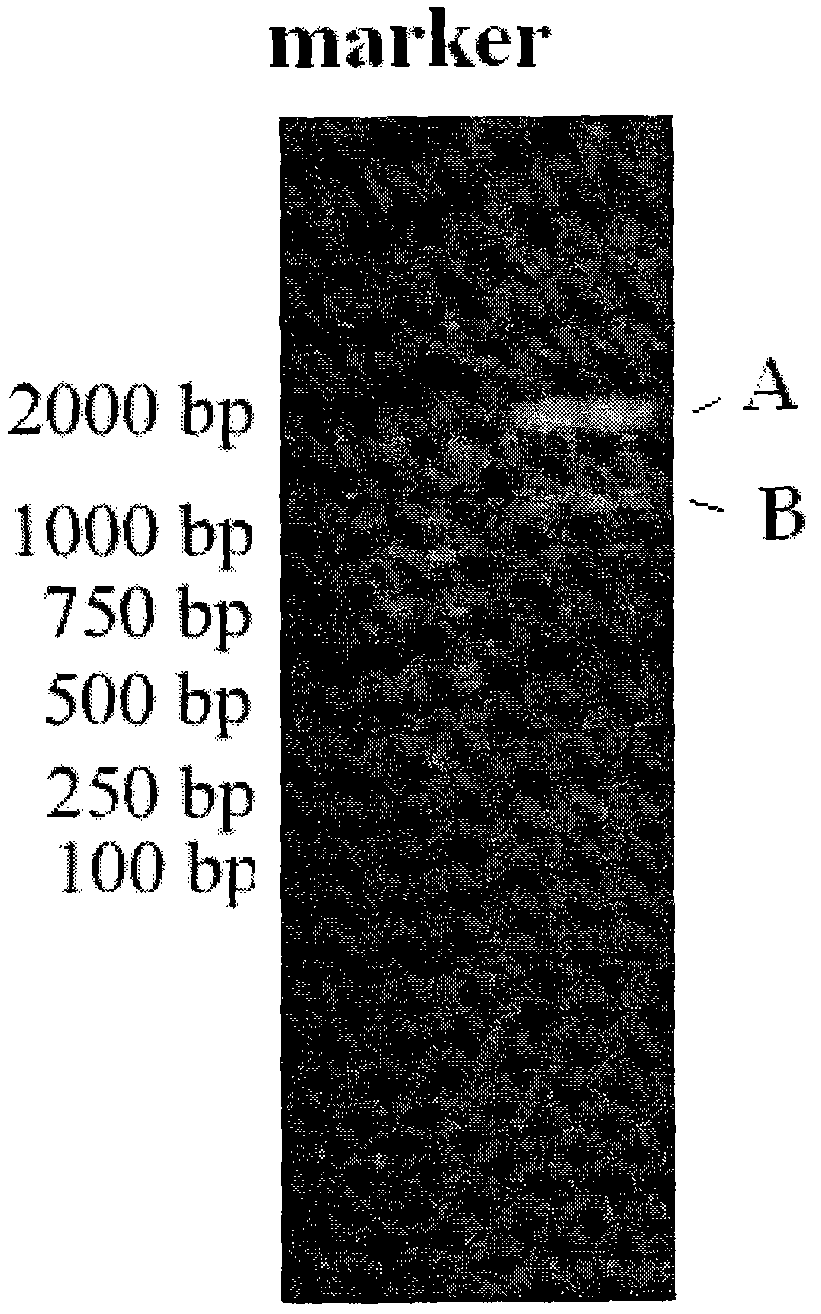
[0024] After double digestion with BamHI and Sac I from the cloning vector plasmid containing the preferred codon-optimized dihydroxybiphenyl dioxygenase gene, the DNA recovery kit recovers about 900 bp of the dihydroxybiphenyl dioxygenase gene fragment. The fragment was connected with the corresponding enzyme-cut vector, and the correct Pseudomonas fluorescens expression gene vector pFB1690 was obtained after identifi...
Embodiment 2
[0028] Genetic transformation of Pseudomonas fluorescens expressing gene vector
[0029] (1) Test method:
[0030] 1. Competent state preparation
[0031] (1) Streak culture on a 2YT plate at a culture temperature of 29°C.
[0032] (2) Pick a single colony of fresh Agrobacterium with a diameter of 2-3 mm on the 2YT solid medium, and spread the colony by Vortex shaking in a sterile eppendorf containing 1 mL of sterile 2YT culture medium.
[0033] (3) In 5 mL of 2YT culture medium, culture overnight at 29° C. and 275 rpm.
[0034] (4) Dilute 5mL of bacterial liquid into 500mL of 2YT liquid culture medium, and cultivate to OD at 29°C 550 0.5~0.7(2~5×10 8 cells / mL).
[0035] (5) Pour the bacterial solution into a pre-cooled centrifuge tube, centrifuge at 2600g for 12-15 minutes at 4°C, and pour off the supernatant carefully.
[0036] (6) Sufficiently resuspend the bacteria with an equal volume of sterile water pre-cooled at 4°C, centrifuge at 3500g for 10 minutes, and pour o...
Embodiment 3
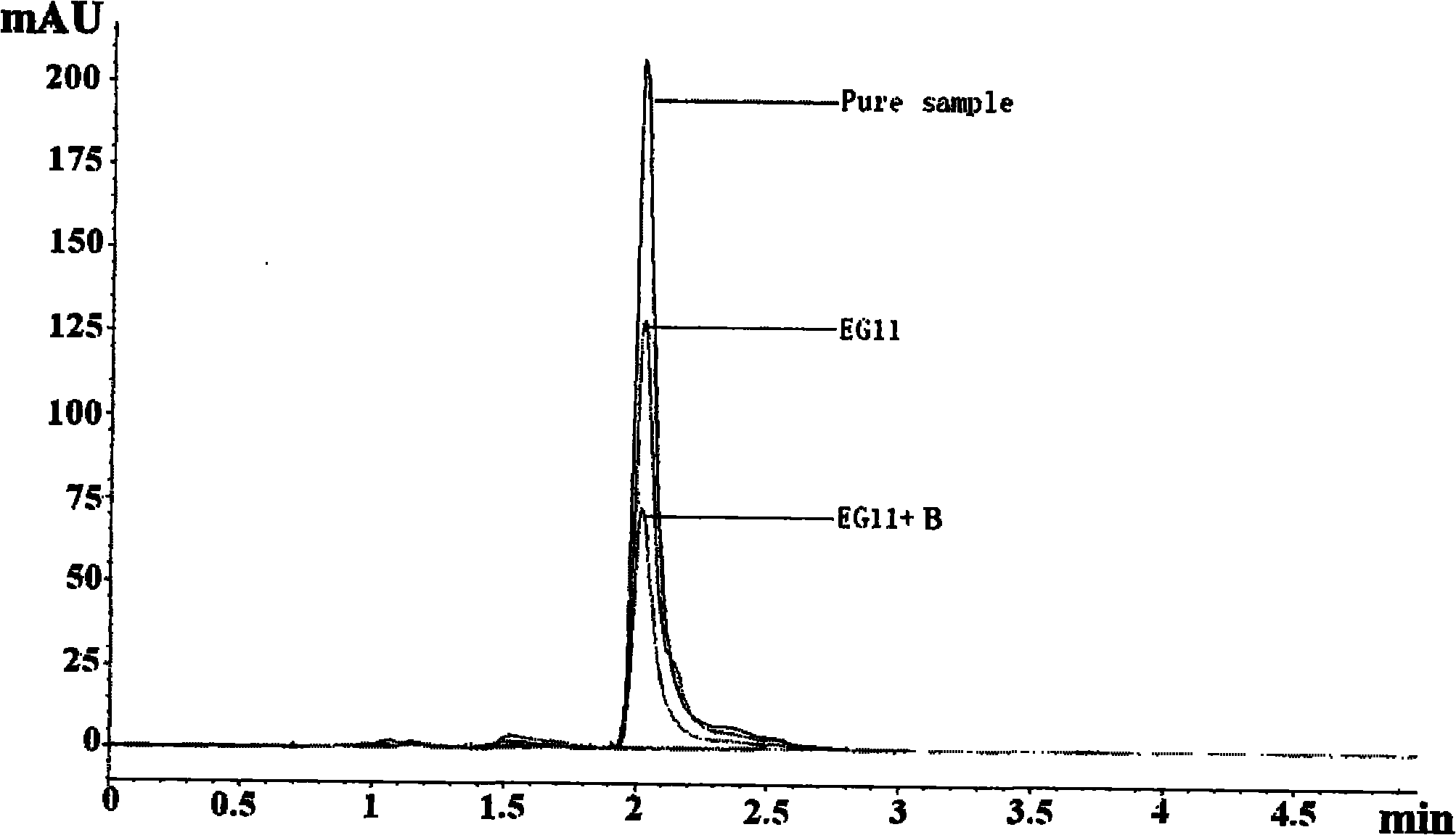
[0049] HPLC Determination of Degradation of Dihydroxybiphenyl
[0050] (1) Test method:
[0051] Steps for detecting bacterial degradation of biphenyl by HPLC:
[0052] (1) Add 100 mL of LB culture medium to a 200 mL Erlenmeyer flask, add 100 μL of the strain to be selected, and cultivate overnight at 28°C on a shaker at 120 rpm.
[0053] (2) On the second day, when the bacteria grow well, draw 1 mL and add 10 μL of dihydroxybiphenyl (dissolved in 0.5 μg / μL methanol) prepared in advance. Let stand at room temperature for 2 minutes.
[0054] (3) Pipette 500 μL of bacterial liquid into Dorf tubes (2 tubes, namely 1000 μL), add 500 μL of ether respectively, and mix well (it is recommended to vibrate for 5-10 minutes in an oscillator), so that dihydroxybiphenyl can be fully extracted by ether , aspirated the supernatant, and placed in a new Dorf tube (combined into one tube).
[0055] (4) Finally, add 500 μL of methanol into the Dorf tube, mix well, and perform rotary evaporatio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com