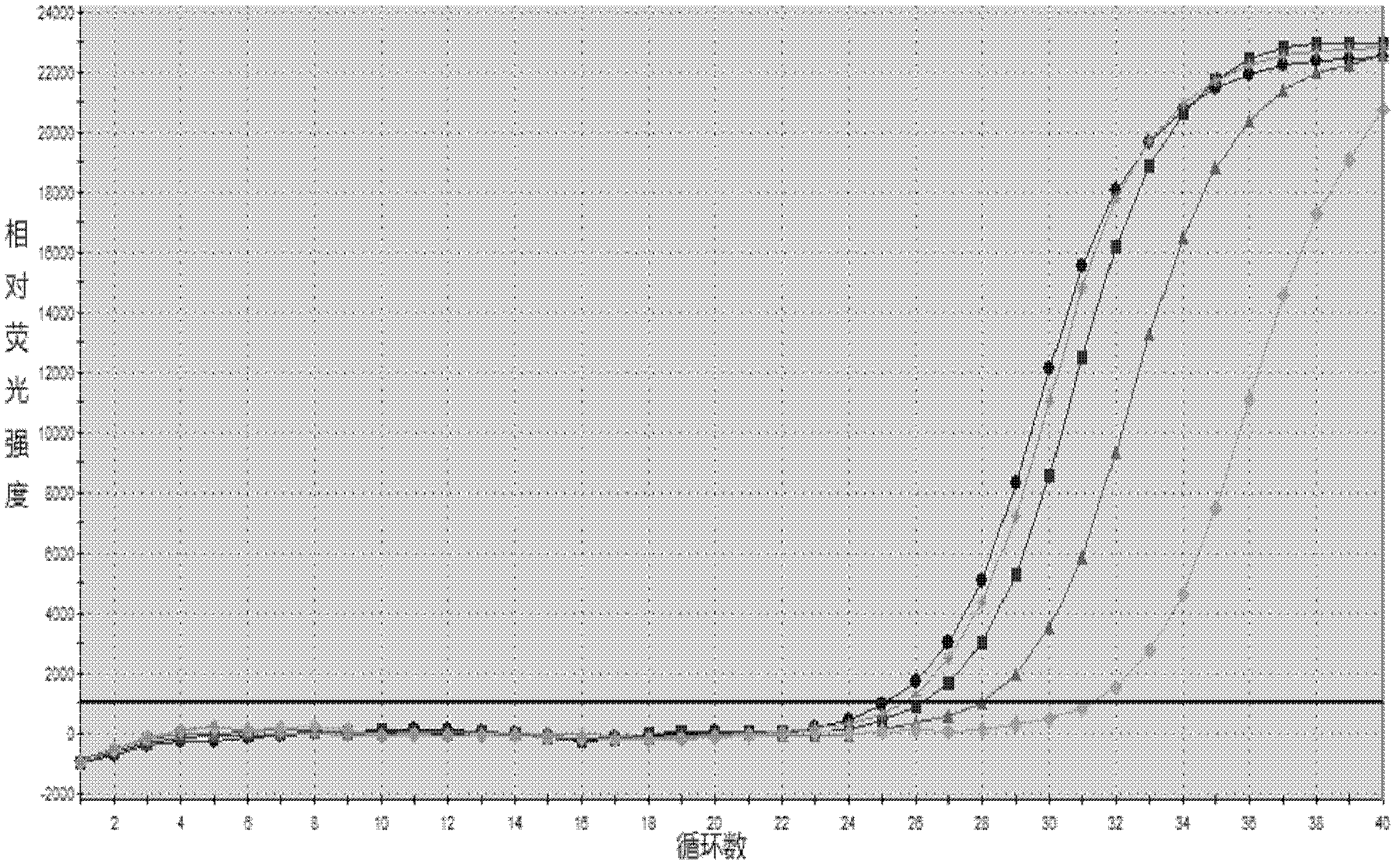PCR (polymerase chain reaction) fluorescence quantitative rapid test kit and method for chlamydia trachomatis
A Chlamydia trachomatis and detection kit technology, which is applied in the direction of microbial measurement/inspection, fluorescence/phosphorescence, biochemical equipment and methods, etc., can solve the problems of cumbersome operation and operational pollution, and achieve quantitative accuracy, high accuracy, and sensitivity and the effect of precision improvement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Composition and configuration of embodiment 1 kit
[0042] 1. Main raw material sources and preparation methods
[0043] The CT primers and probe sequences of this kit are as follows:
[0044] Upstream primer: 5′-CTGTACATTAGGAGCCACCA-3′;
[0045] Downstream primer: 5′-CAACAGATTGATCAAAGTC-3′;
[0046] Fluorescent probes:
[0047] 5'-FAM-GATATCTTAAAGGAAATTCAGCATCTTTCAA-TAMRA-3'.
[0048] The internal reference (Suc) primer upstream primer sequence of this kit: SEQ ID NO.4, downstream primer sequence: SEQ ID NO.5 and probe sequence: SEQ ID NO.6, specifically as follows:
[0049] Upstream primer: 5′-TCATCGTCGCTGGAGCTGGTT-3′;
[0050] Downstream primer: 5′-CGGCGGTTTGTCAAGCTGAT-3′;
[0051] Fluorescent probes:
[0052] 5'-HEX-CTTCTTATAGTCACTGCACTAAACTGGAT-TAMRA-3'.
[0053] The kit also includes DNA polymerase, strong positive quality control substance, weak positive quality control substance, negative quality control substance, positive quantitative reference substan...
Embodiment 2
[0094] The use of embodiment 2 kit
[0095] a. Reagent preparation (reagent preparation area)
[0096] 1. Take out the DNA extraction solution and set aside.
[0097] 2. After determining the number of reaction tubes n (number of samples + negative + positive quality control + weak positive quality control), take out the CT-PCR reaction solution and absorb n×44μl CT-PCR reaction solution, n×1μl DNA polymerization For the enzyme system, add to a centrifuge tube and vortex to mix well. After brief centrifugation, divide into n PCR reaction tubes, 45 μl per tube. Cover the tube cap and transfer to the sample loading area, and store in a 4°C refrigerator in the dark for future use.
[0098] 3. Transfer the quality control substance and quantitative reference substance to the sample processing area, and put them in a 4°C refrigerator for later use.
[0099] b. Sample processing (sample processing area)
[0100] sample requirements
[0101] 1. Applicable sample types: reproducti...
Embodiment 3
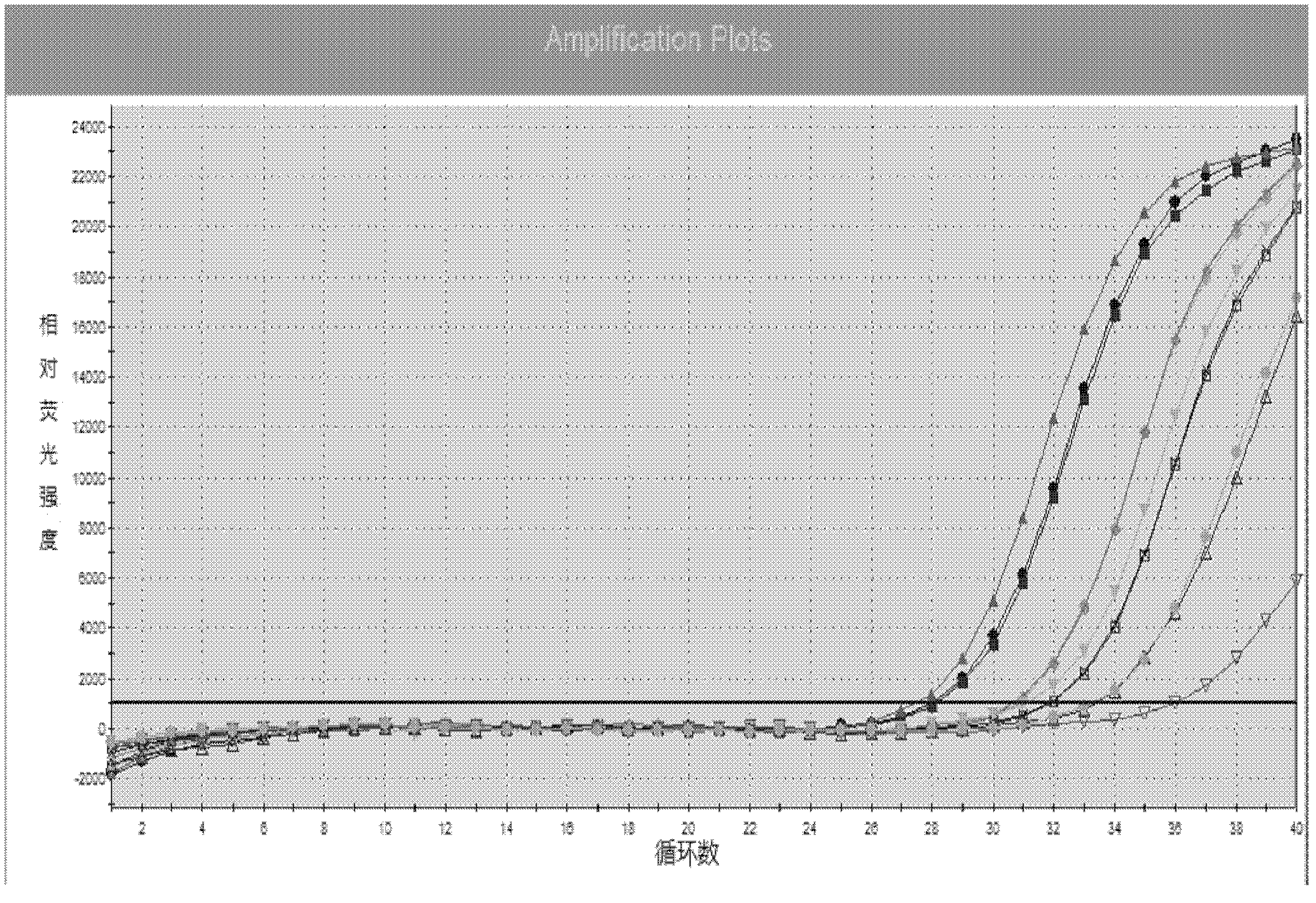
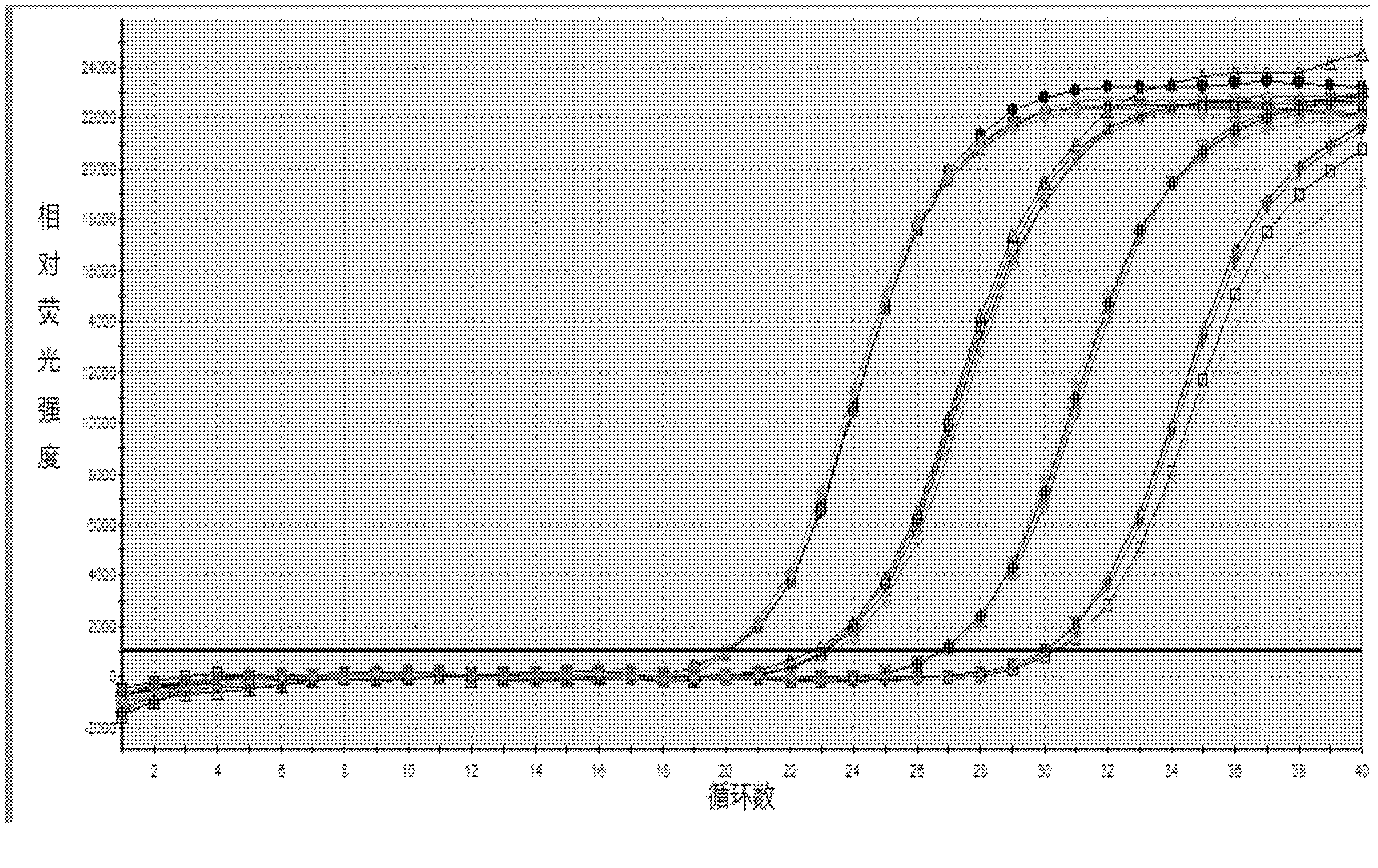
[0145] Embodiment 3 This kit performance evaluation
[0146] The DNA concentration of the positive quantitative reference product is P1: 1.0×10 7 copies / ml, P2: 1.0×10 6 copies / ml, P3: 1.0×10 5 copies / ml, P4: 1.0×10 4 copies / ml.
PUM
| Property | Measurement | Unit |
|---|---|---|
| electrical resistivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com