Unmarked gene knock-out method of extremely acidophilic thiobacillus ferrooxidans
A technology of Thiobacillus ferrooxidans and marker-free genes, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganisms, etc., can solve the problems of less antibiotic availability, antibiotic resistance gene residues, and inconvenient multi-gene knockout Research and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
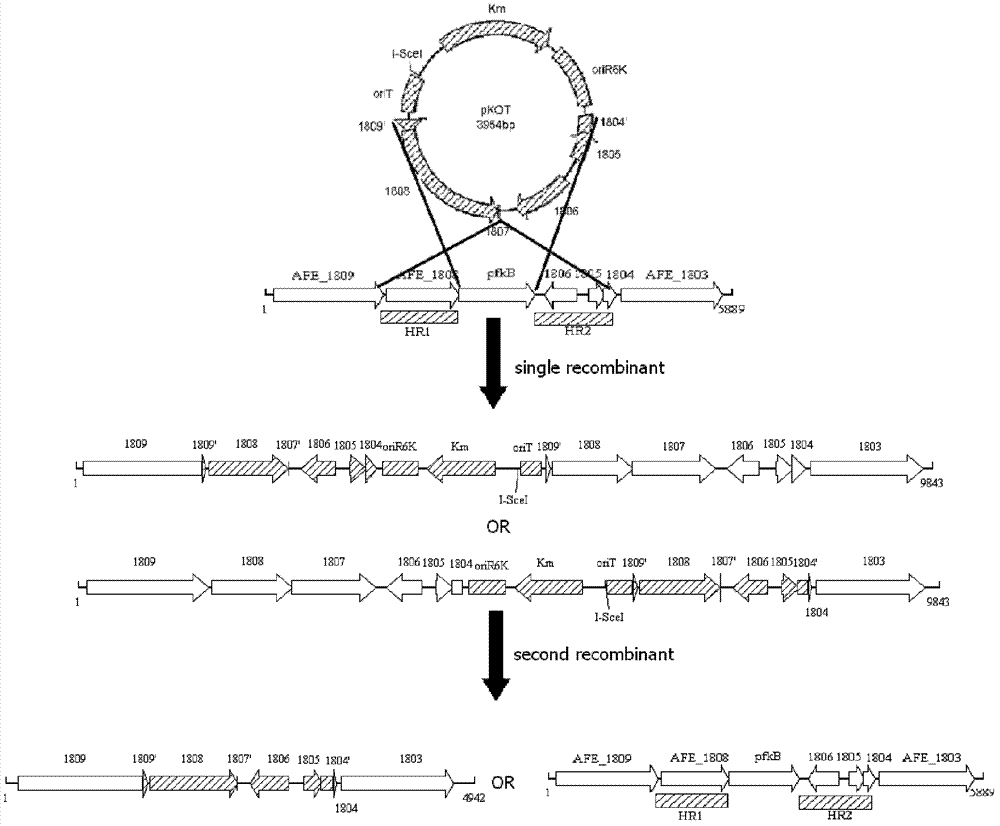
[0040] Example 1: Unmarked knockout of the extreme acidophilic Thiobacillus ferrooxidans phosphofructokinase gene (pfkB, AFE_1807 gene)
[0041] (1) Construction of a suicide plasmid for knocking out the A.ferrooxidans ATCC 23270AFE_1807 gene
[0042] 1. Construction of the initial plasmid for A.ferrooxidans gene knockout
[0043] Primers were designed according to the sequence information of plasmid pSW29T published in GenBank: AY733073.1:
[0044] 29TKpnI:
[0045] 5′-CGGC GGTACCTAGGGATAACAGGGTAAT AGCGCTTTTCCGCTGCATAACCC-3′
[0046] 29TSalI:
[0047] 5′-CTAA GTCGACTGATCAACGCGTCTCGAG GCCGGCCAGCCTCGCAGAGCAGG-3′
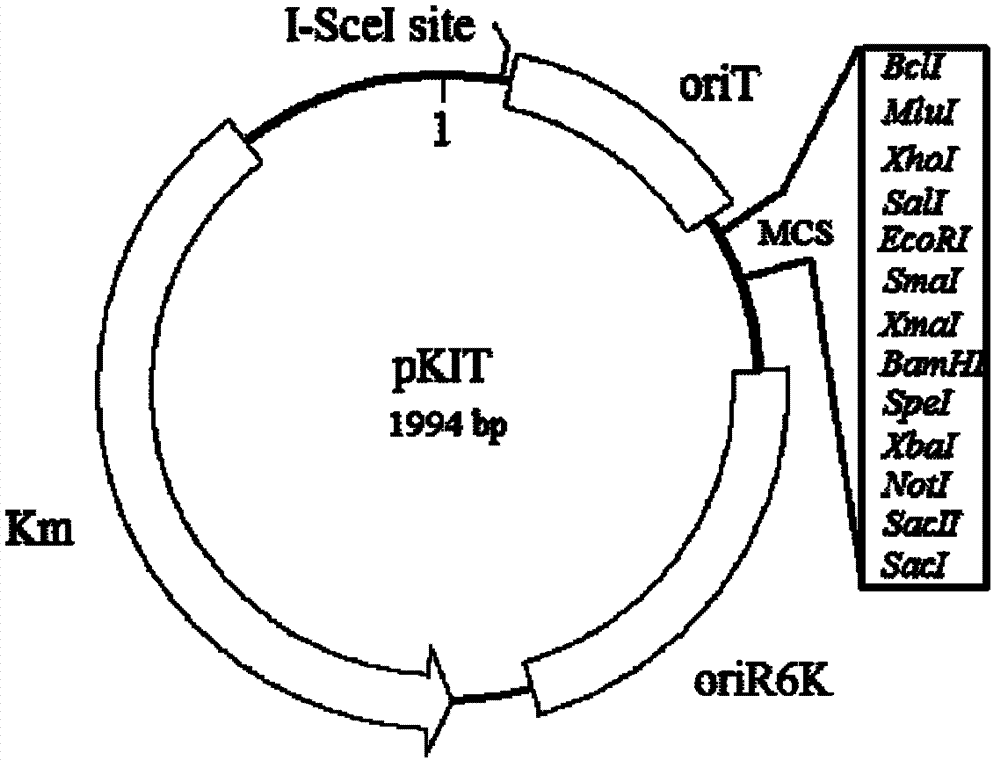
[0048] The 29TKpnI and 29TSalI primers use the plasmid pSW29T as a template to amplify in vitro the fragment with the oriT region by PCR (polymerase chain reaction). The I-SceI recognition and action site sequence of 18 bp is added inside the Kpn I site of the upstream primer, and the Bcl I, Mlu I and Xho I recognition site sequences are added inside the Sal ...
Embodiment 2
[0117] Example 2: Unmarked knockout of the extreme acidophilic Thiobacillus ferrooxidans tetrathionite hydrolase gene (tetH, AFE_0029 gene)
[0118] (1) Construction of a suicide plasmid for knocking out the A. ferrooxidans ATCC 23270 AFE_0029 gene
[0119] 1. construction is used for the initial plasmid of A.ferrooxidans gene knockout (same as embodiment 1, omitted)
[0120] 2. Construction of a suicide plasmid for knocking out the AFE_0029 gene
[0121] Primers were designed according to the AFE_0029 gene and its upstream and downstream gene sequences in the genome sequence of A. ferrooxidans standard strain ATCC 23270 published in GenBank: CP001212.1:
[0122] HA1F:
[0123] 5′-GCT GAATTC CTCGTCCATCATTTTTTTGCGG-3′
[0124] HA1R:
[0125] 5′-TACAAGCTTC TCTAGA CAGATGGTGCGTTTTCC-3'
[0126] HA2F:
[0127] 5′-CGCGAATTC TCTAGA CCGTCTACCTGACCAACTC-3′
[0128] HA2R:
[0129] 5′-GCTAAGCTT GAGCTC CGTCACGTCACTGCGAAAT-3′
[0130] HA1F and HA1R primers, HA2F and HA2R p...
Embodiment 3
[0169] Example 3: Marker-free knockout of the metallo-beta-lactamase gene (AFE 0269 gene, abbreviated as sdo) of Thiobacillus ferrooxidans extreme acidophilus
[0170] (1) Construction of suicide plasmid for knocking out A. ferrooxidans ATCC 23270 AFE_0269 gene
[0171] 1. Construction of the initial plasmid for A. ferrooxidans gene knockout (same as Example 1, omitted)
[0172] 2. Construction of suicide plasmid for knocking out AFE_0269 gene
[0173] Primers were designed according to the AFE_0269 gene and its upstream and downstream gene sequences in the genome sequence of A. ferrooxidans standard strain ATCC 23270 published by GenBank: CP001212.1:
[0174] HB1F:
[0175] 5′-CAC GAATTC CACGGACTGGGCGGATTAT-3′
[0176] HB1R:
[0177] 5′-CAC TCTAGA ATCACCCCTTGTCGGCAATG-3′
[0178] HB2F:
[0179] 5′-CCG TCTAGA GAGGAGTACTTAAGATGAGCAAACA-3′
[0180] HB2R:
[0181] 5′-GAT GAGCTC TGGTCGGTGATGACGGTACGG-3′
[0182] The HB1F and HB1R primers, HB2F and HB2R primers, re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com