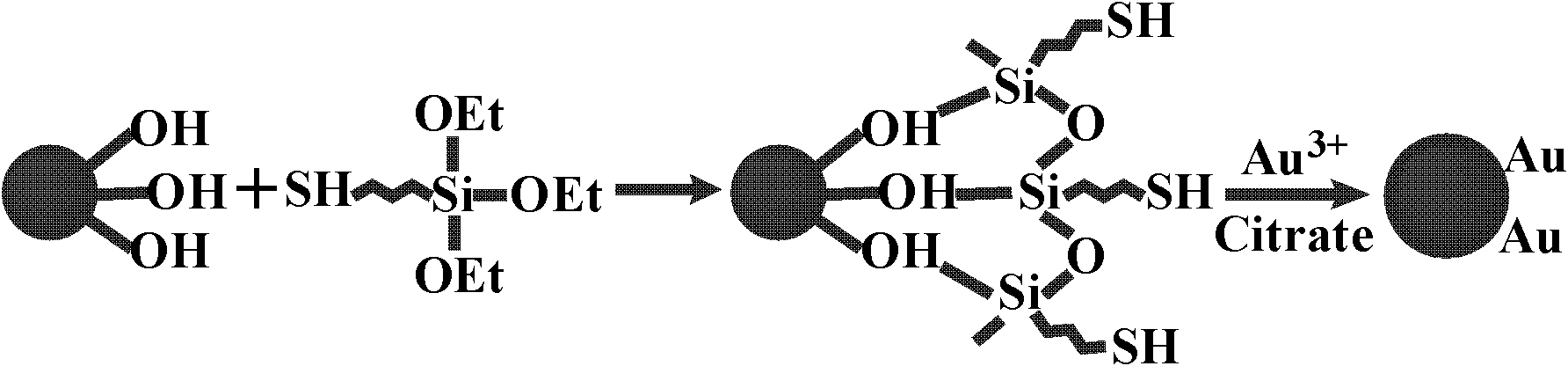Nucleotide sequence analysis method based on magnetic separation and high-fidelity polymerase correction function
A high-fidelity polymerase and nucleic acid sequence analysis technology, which is applied in biochemical equipment and methods, and the determination/inspection of microorganisms. It can solve the problems of insufficient read length and low background signal, so as to reduce background noise and signal accumulation. effect, the effect of great application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Preparation of gold magnetic nanoparticles suitable for sequencing
[0037] Magnetic nanoparticles refer to materials with at least one dimension between 1-1000 nm in particle size. When they reach a certain critical size, they will have a single magnetic domain and exhibit superparamagnetism. It has been gradually produced and developed since the 1970s. New biomedical functional materials developed. Under normal circumstances, the magnetic particles in the colloidal state are in a disorderly state of motion, can be stably suspended in the liquid medium, and conveniently carry out various biological reactions; and under the action of an external magnetic field, the magnetic particles show good magnetic properties. Responsiveness, the particles can be easily separated from the medium; when the external magnetic field is removed, the magnetic particles can be resuspended in the liquid medium without aggregation. In the embodiment of the present invention, the f...
Embodiment 2
[0040]Example 2 Verification of ddNTP correction function by Pfu high-fidelity DNA polymerase
[0041] The 3'-5' exonuclease activity is an intrinsic function of many DNA polymerases. It checks the incorporated nucleotides during DNA synthesis and removes nucleotides that do not match the template from the 3'-5' direction. Nucleotides are called the proofreading function of DNA polymerase. According to the different needs of DNA synthesis, various DNA polymerases have been transformed by modern genetic engineering methods, and DNA polymerases with different functions have been obtained. Pfu high-fidelity DNA polymerase is one of them. Pfu high-fidelity DNA polymerase can not only remove nucleotides that do not match the template incorporated during DNA synthesis from the 3'-5' direction, but also remove hindered nucleotides incorporated during DNA synthesis A chain-continuing nucleotide, such as a modified nucleotide with steric hindrance or a ddNTP that terminates the elonga...
Embodiment 3
[0043] Example 3 Sequencing of p-GEM-T Easy plasmid vector based on the principle of nucleic acid sequence analysis based on magnetic separation and high-fidelity polymerase correction function
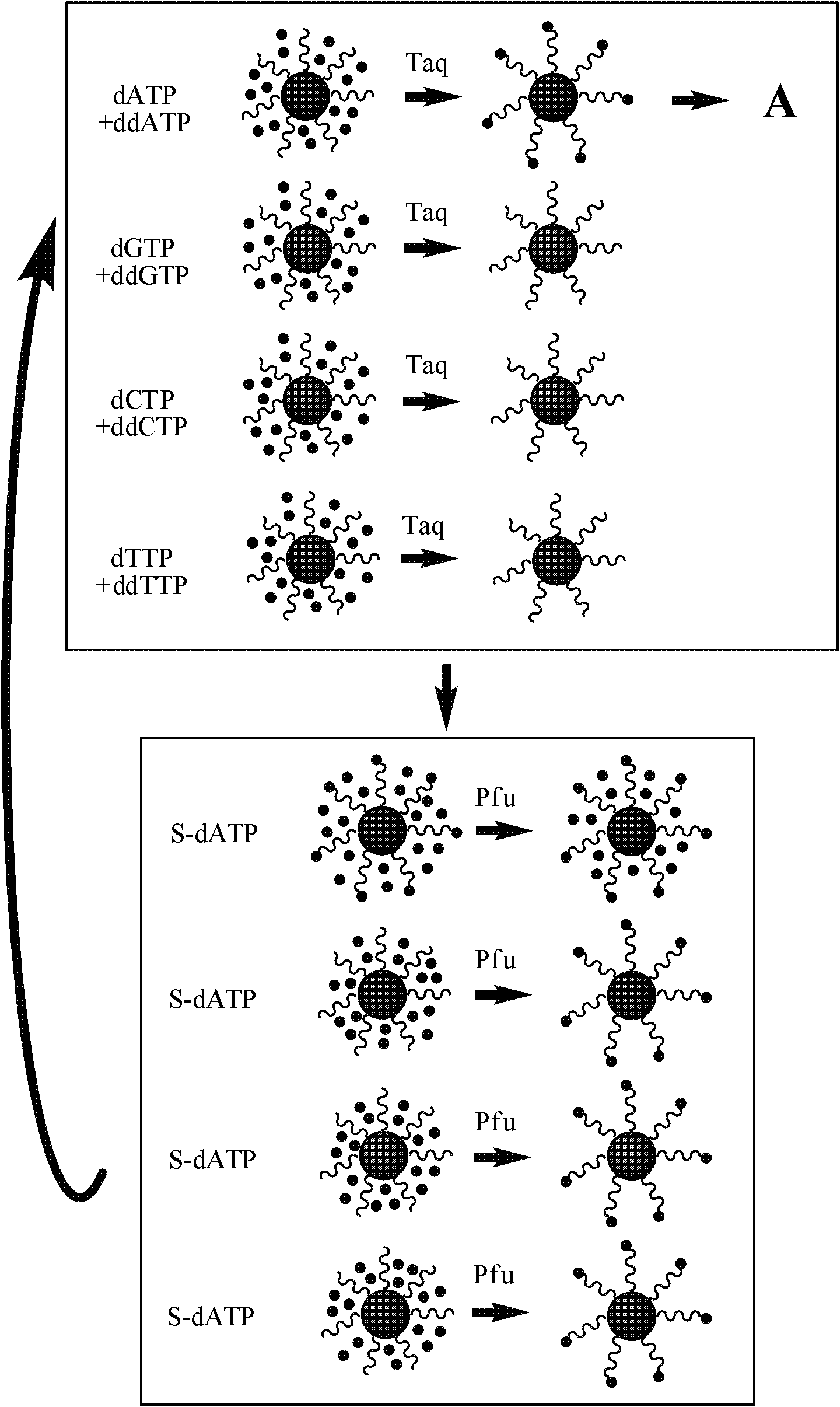
[0044] By modifying streptavidin on the gold magnetic nanoparticles obtained in Example 1, the following sequencing process can be performed:
[0045] ① Design four primers, the first base downstream of which is one of the known A, T, C, and G, and the 5' end is modified with biotin, and the primers are fixed on the Streptavidin-gold magnetic nanoparticles. Cy5-labeled dideoxyribonucleotide monomers ddATP, ddTTP, ddCTP, and ddGTP were divided into four groups for single-base extension reactions in parallel, and the signal values were recorded after scanning with a scanner equipped with Cy5 color filters as follow-up signals Processing benchmarks.
[0046] ② According to the SP6 site of the p-GEM-T Easy plasmid vector, the sequencing primer was designed as follows: TTCTATAGTGTCACCT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com