Nucleic acid aptamer of protein tyrosine phosphatase SHP2 and preparation method thereof
A technology of tyrosine phosphatase and nucleic acid aptamer, which is applied in the field of nucleic acid, can solve the problems that it is difficult to become a drug with therapeutic value, the cell permeability and bioavailability selectivity are not very ideal, and the compound is easy to ionize, etc., to achieve screening The effect of simple and fast detection, easy synthesis and labeling, and small molecular weight
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 In vitro screening of nucleic acid aptamers specifically binding to protein tyrosine phosphatase SHP2
[0040] 1) Dissolve the synthesized 5nmol single-stranded DNA nucleic acid library in binding buffer (12mmol / L PBS, 0.55mmol / LMgCl 2 ), conduct heat treatment: heat at 95°C for 5 minutes, place on ice for 10 minutes, and then place at room temperature for 10 minutes;
[0041] 2) Incubate the processed single-stranded DNA nucleic acid library with GST microbeads, and collect the liquid that is not bound to the GST microbeads;
[0042] 3) Incubate the liquid not bound to the GST microbeads with the SHP2-GST microbeads at 37°C for 40min;
[0043] 4) washing the incubated SHP2-GST microbeads with a binding buffer, and performing a PCR reaction on the SHP2-GST microbeads bound to the oligonucleotide;
[0044] The PCR reaction program was: pre-denaturation at 94°C for 3min, 30s at 94°C, 30s at 53°C, 30s at 68°C, 10 cycles of amplification, and final extension at ...
Embodiment 2
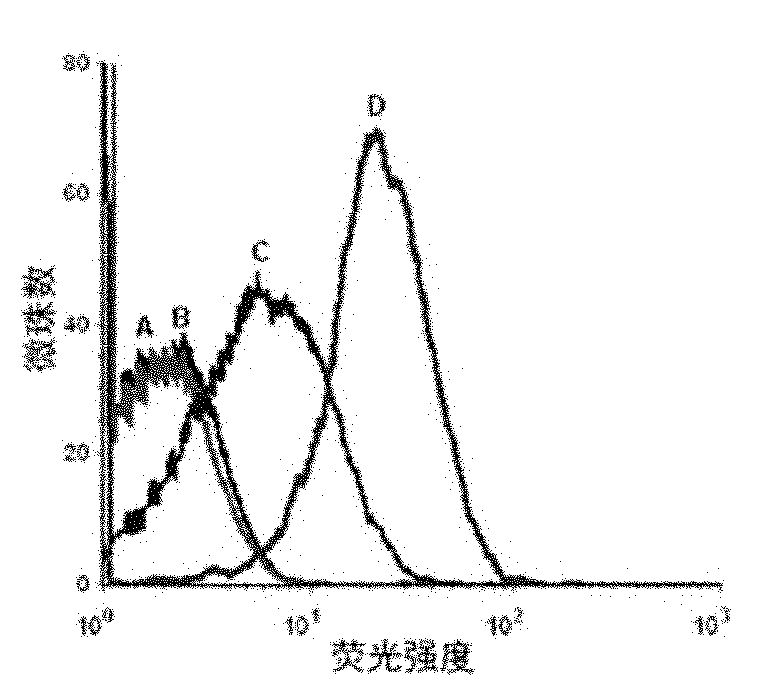
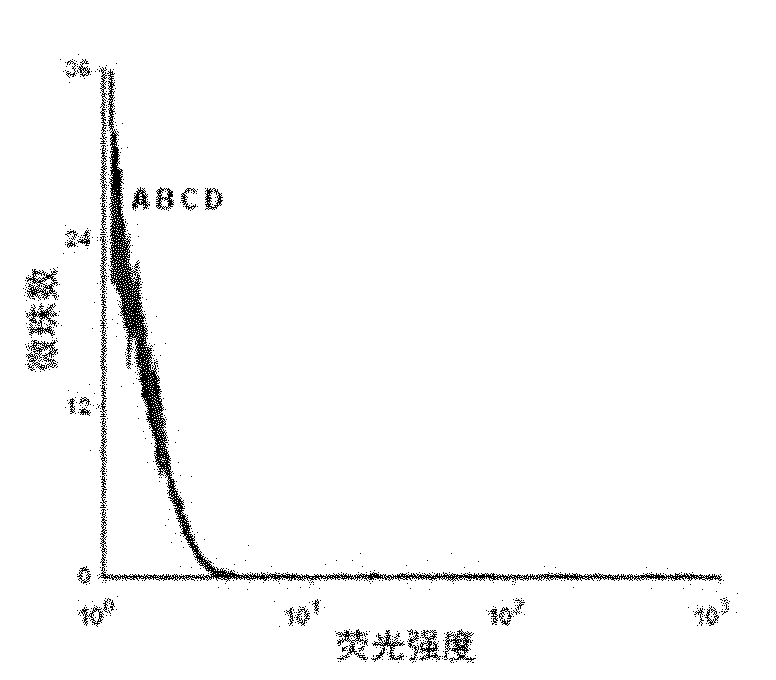
[0049] Example 2 Detection of the binding ability of the resulting single-stranded DNA to the protein tyrosine phosphatase SHP2 by flow cytometry
[0050] First PCR amplifies fluorescently labeled single-stranded DNA, using primer 2: 5′-Biotin-CTGACC ACGAGC TCCATT AG-3′ and primer 3: 5′-FAM-AGC GTC GAA TAC CAC TAC AG-3′, the PCR product For double-stranded DNA with FAM at the 5' end and biotin at the 3' end, add streptavidin microbeads, react for 30 minutes, then use 0.1mol / L NaOH to single-stranded, and purify through a desalting column to obtain FAM-labeled single-stranded DNA for flow cytometry analysis.
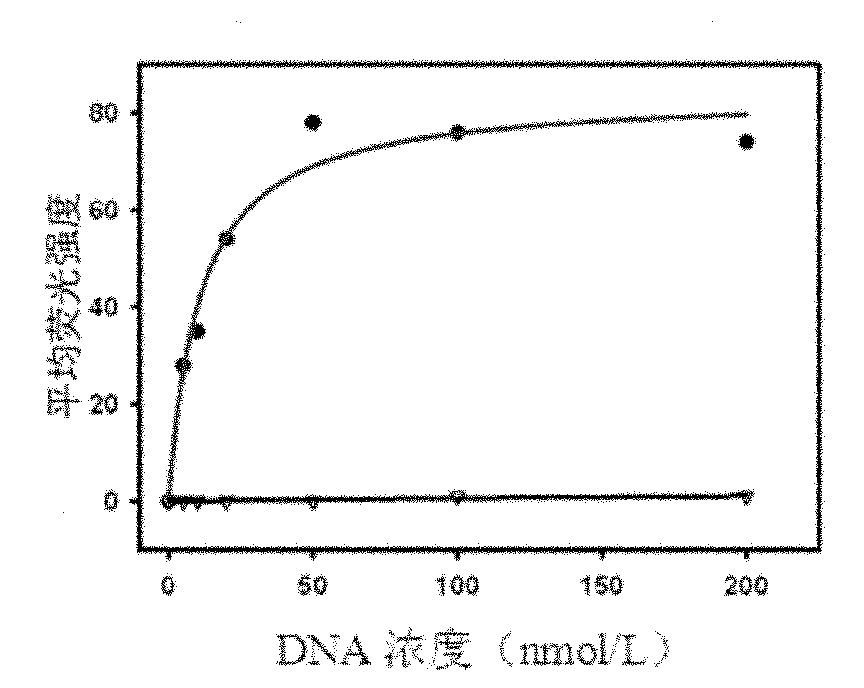
[0051] Use 0nmol / L, 5nmol / L, 10nmol / L, 20nmol / L, 50nmol / L, 100nmol / L, 200nmol / L single-stranded DNA and target protein SHP2-GST beads to determine the dissociation constant (Kd) . Use 200 μl of binding buffer to prepare DNA solutions of the above concentrations, heat at 95°C for 5 minutes, place on ice for 10 minutes, and then place at room temperature for 10 minutes. ...
Embodiment 3
[0053] Example 3 Determination of the obtained nucleic acid aptamer HJ24 by circular dichroism to form a parallel G tetramer structure
[0054] Prepare 1 μmol / L nucleic acid aptamer HJ24 solution with binding buffer, and perform heat treatment: heat at 95°C for 5 minutes, place on ice for 10 minutes, and then place at room temperature for 10 minutes. Then use the circular dichroism spectrometer to scan the CD spectrum from 400nm to 200nm with a step length of 0.1nm, repeat the scan 8 times, and the results form a negative peak and a positive peak at 240nm and 260nm respectively. This peak type is consistent with the literature (10, Sattanathan Paramasivan, Iulian Rujan, Philip H.Bolton. Circular dichroism of quadruplex DNAs: Applications to structure, cation effects and ligand binding, 2007, 43: 324-331.) The characteristic peaks of parallel G tetramers reported in coincidence, so it can be judged The obtained nucleic acid aptamer HJ24 has a parallel G tetramer structure (see ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Dissociation constant | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com