Fluorescence in situ hybridization (FISH) method for fish chromosomes
A fluorescence in situ hybridization, chromosome technology, applied in fluorescence/phosphorescence, biochemical equipment and methods, microbial determination/inspection, etc. clear image effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] a. Preparation of chromosome slide specimens
[0022] Use methods such as erythrocyte nuclear volume measurement or flow cytometry to detect the ploidy of the collected loach, and screen the natural diploid. Chromosomal slide specimens were observed under a phase-contrast microscope, and metaphase specimens with clear images and well-dispersed chromosomes were selected and stored in a -80°C refrigerator.
[0023] b. Pretreatment of chromosome slide specimens
[0024] 1) Take out the chromosome slide specimens stored at -80°C and put them in a 65°C incubator for 2 hours to dry;
[0025]2) Soak the dried chromosome slide specimen in 4×SSC solution for 5 minutes, pipette 100 μL of 0.5 mg / ml RNase solution onto the chromosome slide specimen, cover the slide with parafilm, and place The slides were placed in a wet box with 2×SSC solution at the bottom at 37°C for 30min. Operate in groups of two slides;
[0026] 3) Carefully remove the parafilm with fine tweezers, immedia...
Embodiment 2
[0057] The difference from Example 1 is the preparation of chromosome slide specimens: use methods such as erythrocyte nuclear volume measurement or flow cytometry to detect the ploidy of the collected loach, and screen for natural tetraploids. According to the methods of the prior art, use Live gill tissue was used as material, and natural tetraploid chromosome slide specimens were prepared by air-drying method, observed under a phase-contrast microscope, and metaphase specimens with clear images and well-dispersed chromosomes were selected and stored in a -80°C refrigerator.
[0058] Other steps and signal observation are the same as in Example 1.
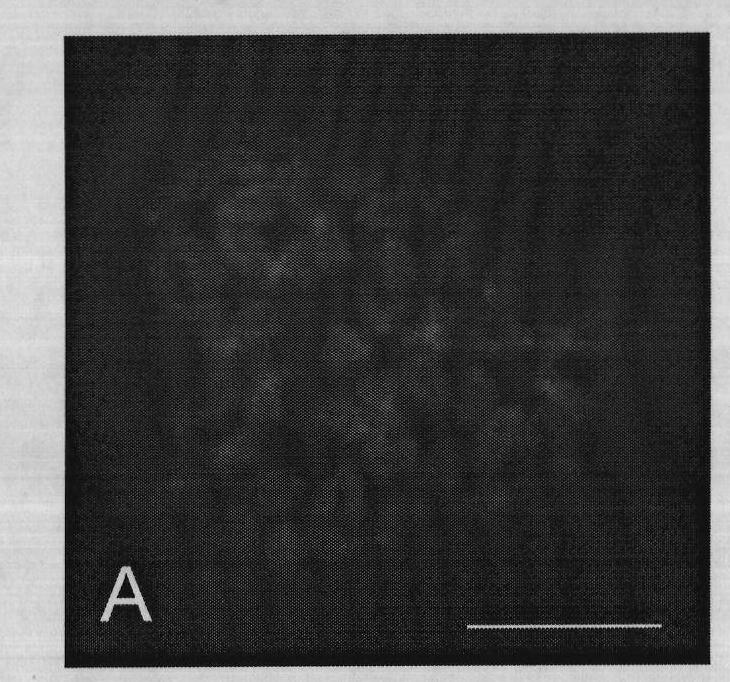
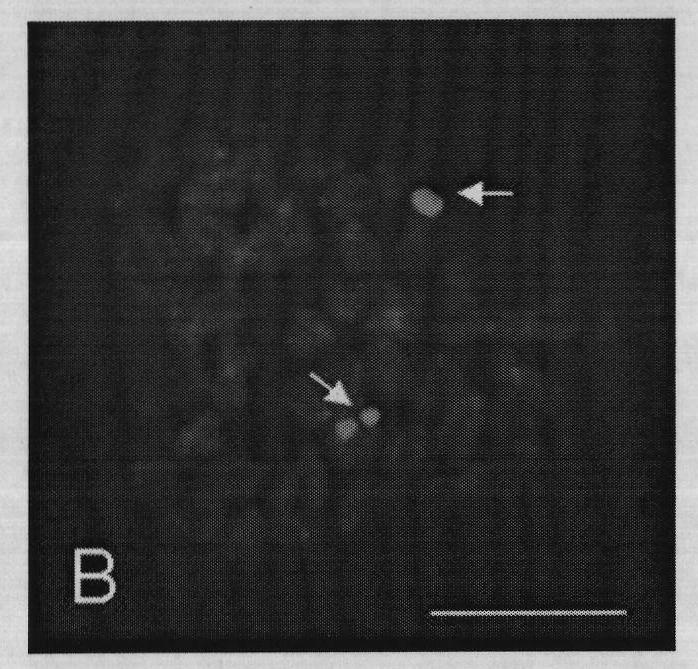
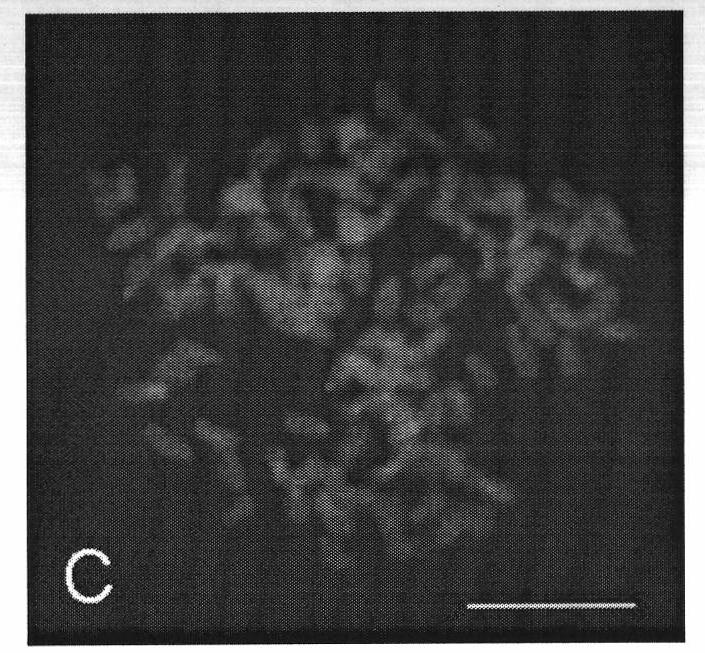
[0059] Metaphase (DAPI counterstaining) effect diagram and fluorescence in situ hybridization effect diagram of metaphase phase are as follows image 3 (C), Figure 4 (D) shown. Chromosomes were counterstained with DAPI, showing blue; arrows indicate hybridization signal sites (green); the scale bar is 10 μm.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com