Kit and method for extracting microbial DNA
A kit and microbial technology, applied in the field of molecular biology, can solve the problems of unsatisfactory extraction of microbial DNA, cumbersome operation steps, and loss of species, and achieve the effects of good versatility, high purity, and fast extraction speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Embodiment 1: the kit for extracting microbial DNA according to the present invention
[0069] The kit for extracting microbial DNA of the present invention comprises:
[0070] Extraction buffer: 120mM guanidine isothiocyanate, 181mM Na at pH 7.0 3 PO 4 . Specific preparation method: Weigh 1.42 grams of guanidine isothiocyanate, add enough distilled water to fully dissolve, then add Na with a pH of 7.0 and a concentration of 1M 3 PO 4 18.1ml, mix thoroughly, and dilute to 100ml.
[0071] Lysis solution I: 100 mM Tris-HCl (pH 8.0), 100 mM EDTA (pH 8.0), 1.5 M NaCl, 1% CTAB, 1% PVP. Specific preparation method: Weigh 1 gram of CTAB and 1 gram of PVP10 respectively, add appropriate amount of distilled water to dissolve respectively, then add 10 ml of 1M Tris-HCl (pH 8.0), 20 ml of 0.5 M EDTA (pH 8.0), 30 ml of 5 M NaCl , and then dilute to 100ml.
[0072] Lysis Solution II: SDS at a final concentration of 2%.
[0073] Inhibitor removal solution I: 250 mM ammonium a...
Embodiment 2
[0079] Embodiment 2: extract microbial DNA from soil
[0080] The kit described in Example 1 was used for extraction.
[0081] 1. Extraction reagent
[0082] Extraction buffer: 120mM guanidine isothiocyanate, 181mM Na at pH 7.0 3 PO 4 .
[0083] Lysis solution I: 100 mM Tris-HCl (pH 8.0), 100 mM EDTA (pH 8.0), 1.5 M NaCl, 1% CTAB, 1% PVP.
[0084] Lysis Solution II: SDS at a final concentration of 2%.
[0085] Inhibitor removal solution I: 250 mM ammonium acetate.
[0086] Inhibitor removal solution II: 120 mM ammonium aluminum sulfate.
[0087] Binding solution: 5M guanidine hydrochloride, 20mM Tris-HCl (pH 7.5), 30% isopropanol.
[0088] Rinsing solution: 10 mM Tris-HCl with a pH value of 7.5, 100 mM NaCl, and 80% absolute ethanol.
[0089] Eluent: 10 mM Tris-HCl, pH 8.0.
[0090] Glass cellulose membrane adsorption column: 0.45 μm glass cellulose membrane, Whatman Company, GF / B type, 1.5 mL spin column.
[0091] 2. Extraction method
[0092] (1) Take the soil sam...
Embodiment 3
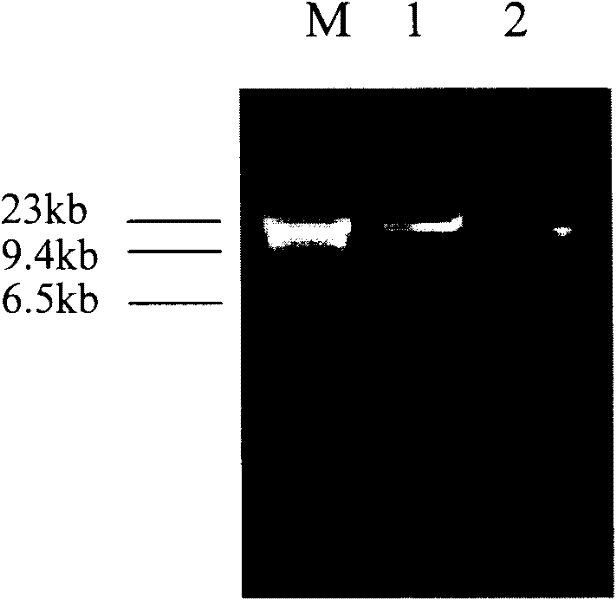
[0100] Embodiment 3: the microbial DNA concentration and purity detection of extraction
[0101] Same as Example 1, the same amount of soil samples were extracted with Mo Bio UltraClean Soil DNA Kit to extract microbial DNA, and the microbial DNA extracted in Example 1 and the microbial DNA extracted with Mo BioUltraClean Soil DNA Kit were tested for concentration and purity. The results are shown in the table 1.
[0102] Table 1 Detection of microbial DNA concentration and purity
[0103] The method of the invention
[0104] The results show that the concentration of microbial DNA extracted in Example 1 is greater than the concentration of microbial DNA extracted with Mo BioUltraClean Soil DNA Kit; The value is closer to the standard value (1.7-2.0) than the DNA extracted by the MO BIO kit. The above results show that with the same sample size, the DNA extracted by the present invention has a high concentration and less impurities.
PUM
| Property | Measurement | Unit |
|---|---|---|
| pore size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com