Acid amylase AMYA4 and gene and application thereof
An acid amylase, gene technology, applied in the field of genetic engineering, can solve the problem of not degrading starch and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0131] Example 1 Cloning of Alicyclobacillus peridum A4 (CGMCC No.3147) amylase-encoding gene AMYA4
[0132] Acquisition of gene sequence
[0133] According to the amylase gene sequence reported by Alicyclobacillus, and with reference to the mass spectrum sequence (GNGQWNIDFFGGDLK, HWLALGADGW) of the above-mentioned purified amylase, specific primers AF and AR were directly designed to use Alicyclobacillus hesperidum A4 (CGMCC No. .3147) Total DNA was used as template for PCR amplification. The PCR reaction parameters were: denaturation at 94°C for 5 min, annealing at 60°C for 30 sec, and extension at 72°C for 1.5 min. Finally, keep warm at 72°C for 10 minutes. A fragment of about 480bp was obtained, which was recovered and connected with the pEASY-T3 vector and sent to Sanbo Biotechnology Co., Ltd. for sequencing.
[0134] According to the nucleotide sequence obtained by sequencing, three TAIL-PCR specific nested primers were designed upstream and downstream respectively: ...
Embodiment 2
[0141] The activity analysis of embodiment 2 purifying amylase
[0142] The method for separating and purifying acid amylase AMYA4 from Alicyclobacillus hesperidum A4 comprises the following steps:
[0143] 1) After shaking the seed medium at 55°C for 48 hours, transfer 1% of the inoculum to the enzyme-producing medium, and after shaking at 55°C and 200rpm for 48 hours, centrifuge the bacterial solution, and take the supernatant for purification;
[0144] 2) The culture was centrifuged to take about 101 of the supernatant, and then passed through a 6K hollow fiber ultrafiltration membrane to about 600ml in an ice bath, and then further concentrated to 200ml with a 5K ultrafiltration membrane bag;
[0145] 3) Remove 10ml of the above-mentioned concentrated solution in buffer A (20mmol / L citric acid-Na 2 HPO 4 pH 3.5) for overnight dialysis, cation exchange chromatography: take 2.0mL of the concentrated solution and put it on the HiTrap SP XL cation column equilibrated with buff...
Embodiment 3
[0147] The property determination of embodiment 3 purifying amylase AMYA4
[0148] 1. The optimum pH and pH stability assay methods of purified amylase AMYA4 are as follows:
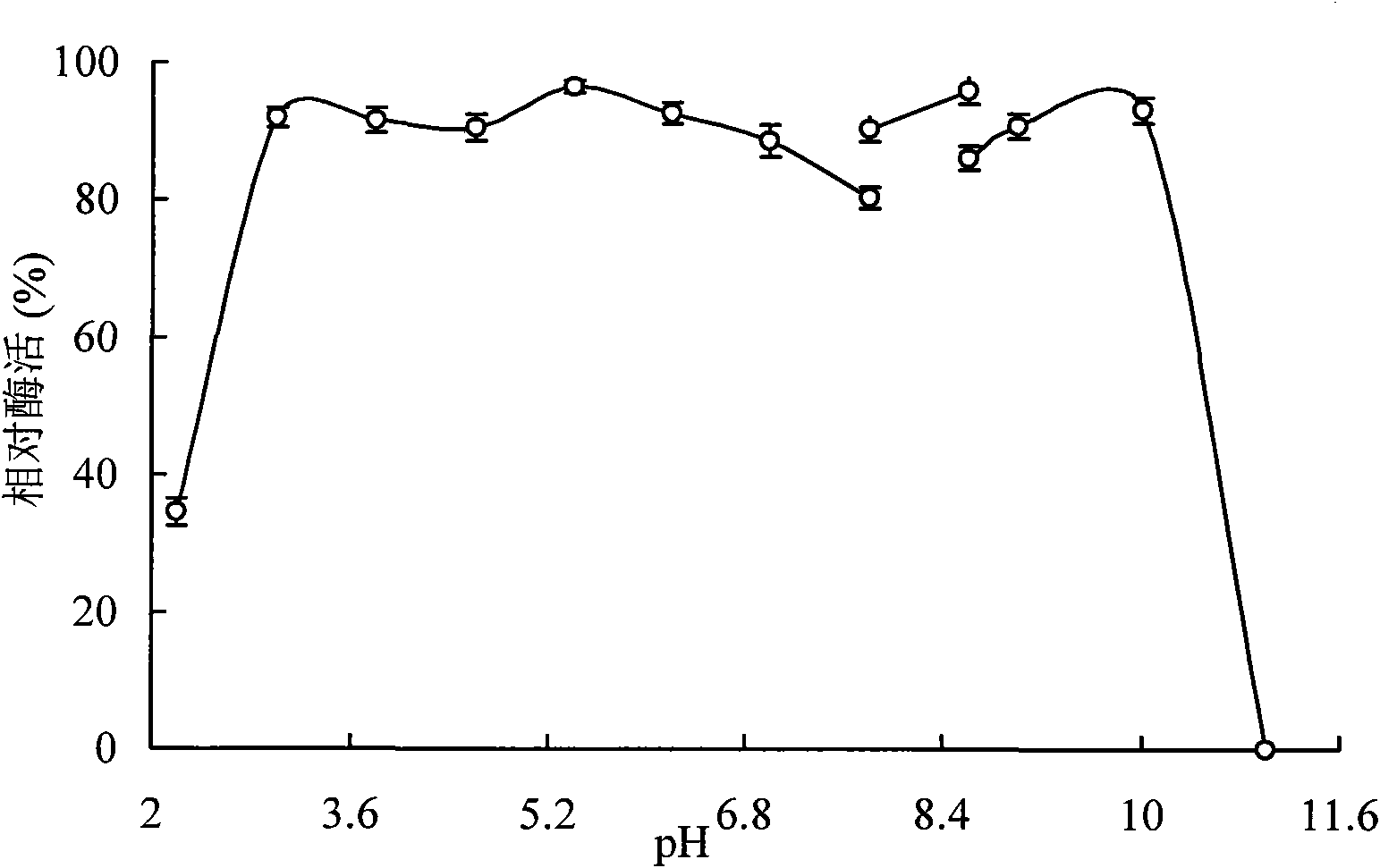
[0149] The amylase purified in Example 2 was subjected to enzymatic reactions at different pHs to determine its optimum pH. Substrate soluble starch with different pH buffers (0.1mol / L citric acid-sodium hydrogen phosphate pH 3.0-7.4; 0.1M Tris-HCl pH 7.8-8.6; 0.1M glycine-sodium hydroxide pH 9.0-11.0;), Amylase activity assays were performed at 75 °C. result( figure 2 ) shows that its optimum pH is 4.2, and it maintains more than 70% relative enzyme activity at pH 3.8-5.8. The most stable pH range of the enzyme is 3.0-10.0, and the enzyme activity is kept above 85% after acting for 2 hours at 37°C. For the experimental results of the optimum action temperature and thermal stability of amylase, see image 3 .
[0150] 2. The optimum temperature and thermostability determination method of amylase i...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com