Human papillomavirus HPV DNA fragment, specific primer and application thereof
A human papillomavirus-specific technology, applied in the field of detection and/or typing of human papillomavirus DNA, can solve problems such as inability to do typing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] Example 1 : Selection of 31 HPV gene DNA fragments
[0079] All the genomes of 31 kinds of HPV viruses published on GenBank were compared, compared for homology, and the 31 kinds of HPV gene DNA fragments provided by the present invention were selected.
[0080]The gene accession numbers of the 31 HPV viruses are as follows (see brackets): HPV6[X00203], HPV 11[M14119], HPV 16[K02718], HPV 18[X05015], HPV 26[X74472], HPV 31[ J04353], HPV 33[M12732], HPV 35[X74477], HPV 39[M62849], HPV 40[X74478], HPV 42[M73236], HPV 43[AJ620205], HPV 44[U31788], HPV 45[X74479] , HPV 51[M62877], HPV 52[X74481], HPV 53[X74482], HPV 54[U37488], HPV 55[U31791], HPV 56[X74483], HPV 58[D90400], HPV 59[X77858], HPV 61[U31793], HPV 66[U31794], HPV 68[X67161], HPV 70[U21941], HPV 72[X94164], HPV 73[X94165], HPV 81[AJ620209], HPV 82[AB027021], and HPV 83[ AF151983].
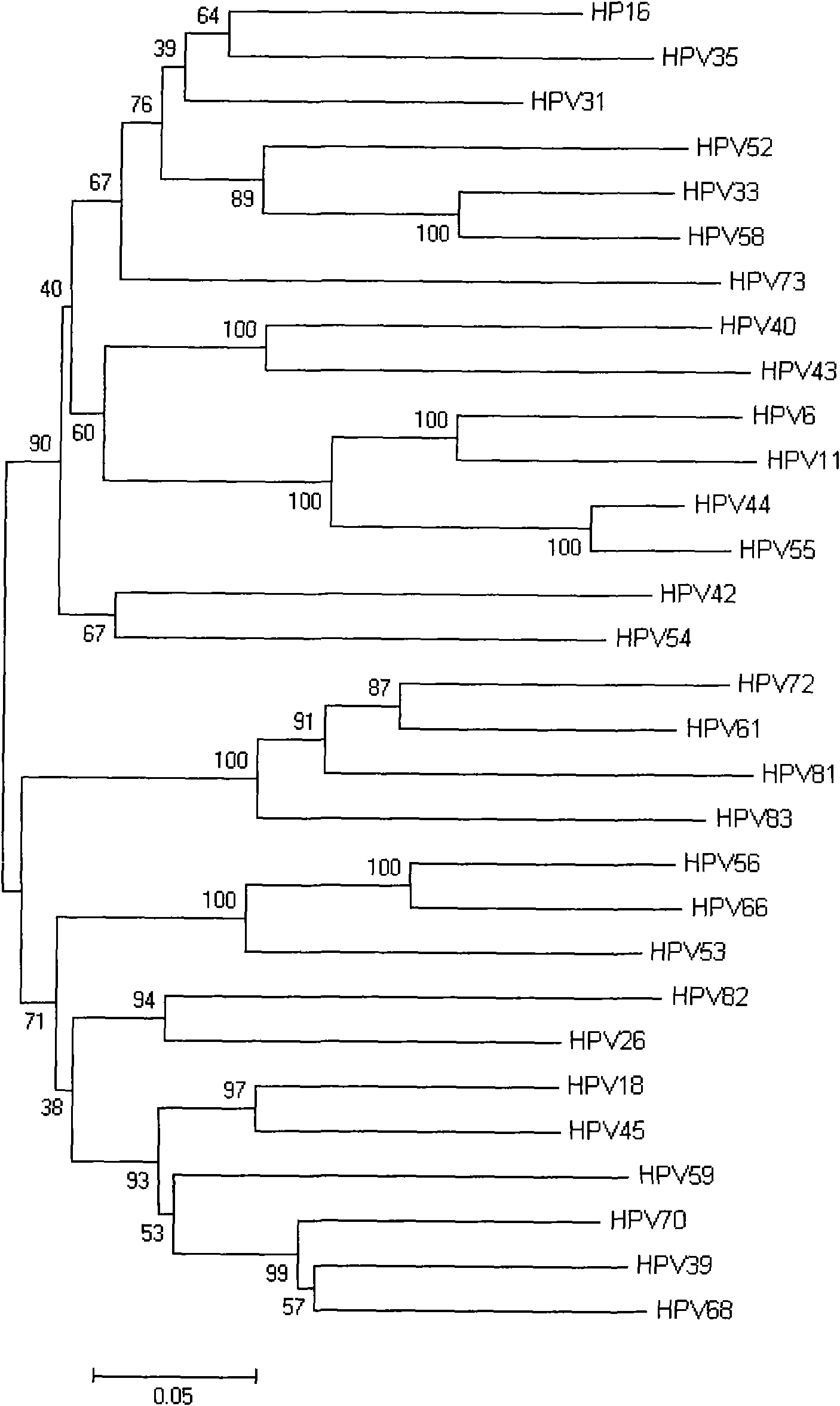
[0081] According to the result of homology comparison, use MEGA4.0 software to make the phylogenetic tree model of each subtyp...
Embodiment 2
[0083] Example 2 : Design multiple pairs of different primers for 31 HPV gene DNA fragments
[0084] Before designing primers, analyze the properties of the target sequence to be tested, and select highly conserved regions with uniform base distribution for primer design. The length of oligonucleotide primers is 15-30bp. The Tm value of the primers is generally controlled at 55-60°C, and the Tm values of the upstream and downstream primers should be as consistent as possible, generally not exceeding 2°C. The proportion of (G+C) in effective primers is generally 40-60%.
[0085] The present invention uses primer software to design and select according to the score of each primer. For each fragment, 10 pairs of primers with the highest scores were selected for testing, and the primers that could not amplify the target band were deleted and re-selected.
[0086] The specific primers of all 31 kinds of human papillomavirus DNA fragments described in Example 1 designed accor...
Embodiment 3
[0118] Example 3 : Amplify 31 HPV DNA templates with 31 pairs of single primers
[0119] Using the 31 pairs of specific primers designed according to Example 2, the corresponding 31 HPV DNA templates were individually amplified by PCR. The primer pairs used are SEQ ID NO: 32-33, SEQ ID NO: 40-41, SEQ ID NO: 48-49, SEQ ID NO: 56-57, SEQ ID NO: 64-65, SEQ ID NO: 72-73 , SEQ ID NO: 80-81, SEQ ID NO: 88-89, SEQ ID NO: 96-97, SEQ ID NO: 104-105, SEQ ID NO: 112-113, SEQ ID NO: 120-121, SEQ ID NO: 120-121, SEQ ID NO: ID NO: 128-129, SEQ ID NO: 136-137, SEQ ID NO: 144-145, SEQ ID NO: 148-149, SEQ ID NO: 156-157, SEQ ID NO: 164-165, SEQ ID NO: 172-173, SEQ ID NO: 180-181, SEQ ID NO: 188-189, SEQ ID NO: 196-197, SEQ ID NO: 204-205, SEQ ID NO: 212-213, SEQ ID NO: 220- 221. SEQ ID NO: 228-229, SEQ ID NO: 236-237, SEQ ID NO: 244-245, SEQ ID NO: 252-253, SEQ ID NO: 262-261 and SEQ ID NO: 268-269.
[0120] 1) Single primer PCR amplification:
[0121] a) Prepare a 25 μL reaction system ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com