Preparation method of recombinant Fas-associated death domain protein and application thereof
A protein and fusion protein technology, applied in the field of genetic engineering biology, can solve problems such as easy aggregation, deletion, and inability to separate and purify FADD proteins, and achieve high-efficiency expression and separation and purification effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
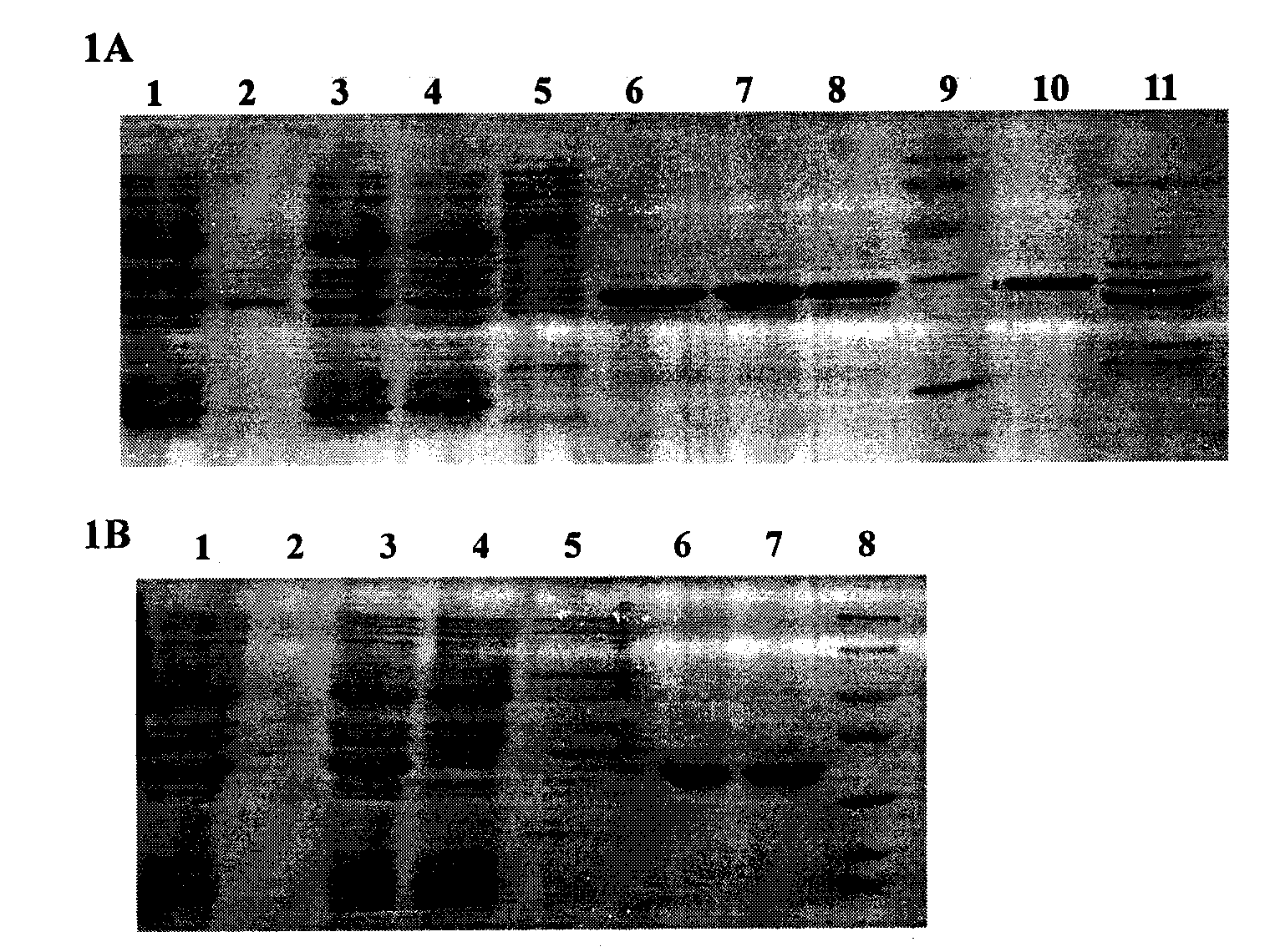
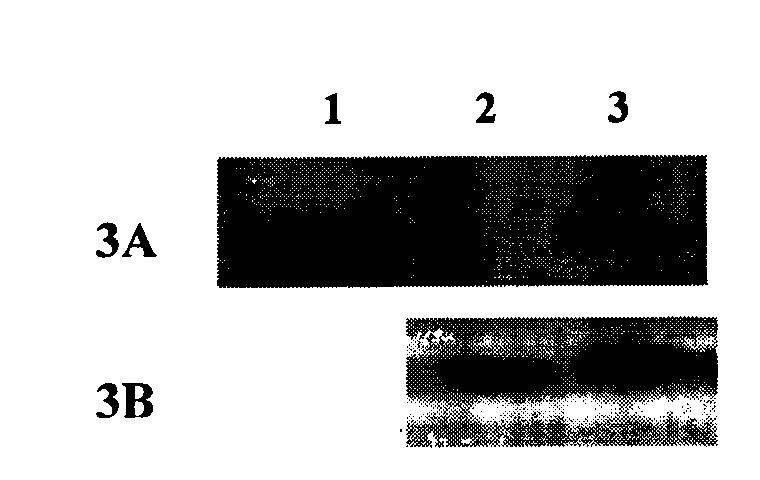
[0034] 1. Construction of His-FADD wild-type protein prokaryotic expression vector:
[0035] According to the gene sequence of FADD, chemically synthesize upstream and downstream primers (upstream primer: 5'-CATGCCATGGGCAGCAGCCATCATCATCATCATCACGGCATGGACCCATTCCTGGTG-3'; downstream primer: 5'-CGCGAAGCTTTCAGGGTGTTTCTGAGG-3'), using the plasmid pGEX2T-FADD previously constructed in our laboratory as a template , the coding sequence of the full-length wild-type FADD was amplified by PCR. The conditions of the PCR reaction were: thermal denaturation at 94°C for 1 minute, annealing at 60°C for 1 minute, and extension at 72°C for 1 minute, and a total of 25 cycles were performed. The PCR product was recovered by agarose gel electrophoresis and subjected to Nco I / Hind III double digestion. Plasmid pET28a (Novagen) was also subjected to Nco I / Hind III double digestion, and the two recovered fragments were digested using T4 DNA ligase. Ligation, the ligation product was transformed into...
Embodiment 2
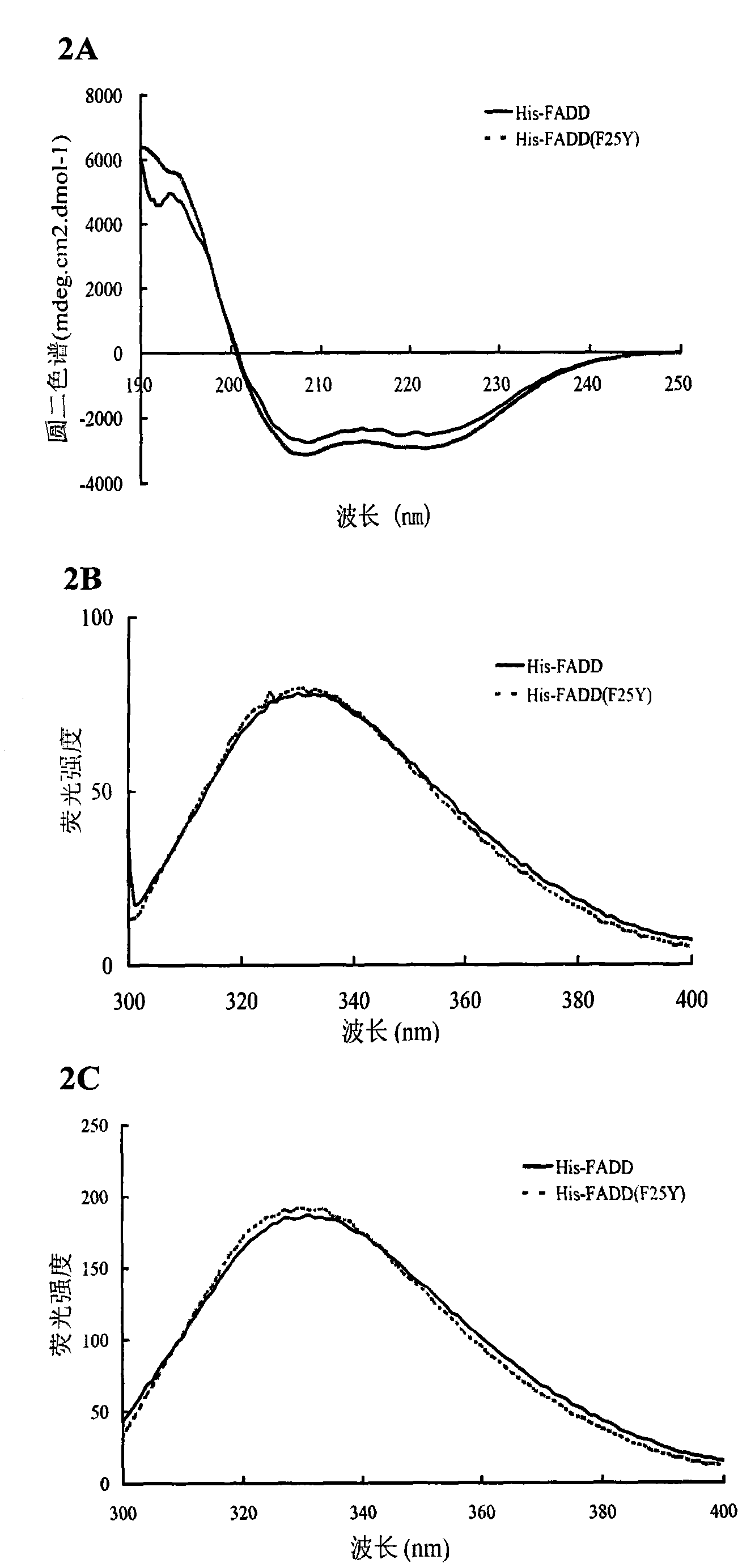
[0057] 1. Construction of wild-type FADD and FADD (F25Y) full-length protein eukaryotic expression vectors
[0058] Using the prokaryotic expression vectors of wild-type FADD and FADD (F25Y) full-length proteins constructed in Example 1 as templates, the coding sequences of wild-type FADD and FADD (F25Y) full-length proteins were amplified by PCR, respectively at the 5' Restriction sites BamH I and Xho I (bold, italic) were introduced at the end and 3' end, and the upstream primer used was: 5'-CGC ATGGACCCATTCCTGGT-3', the downstream primer is: 5'-CCG TCAGGGTGTTTCTGAGG-3'. The conditions of the PCR reaction were: heat denaturation at 94°C for 30 seconds, annealing at 65°C for 30 seconds, and extension at 72°C for 1 minute, for a total of 25 cycles. The purified PCR product was digested with BamH I / Xho I and inserted into the corresponding site of pRK5-FLAG. Perform DNA sequencing on the recombinant expression vector to analyze the correctness of its reading frame and codi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com