Design and selection of genetic targets for sequence resolved organism detection and identification
A sequence and biological technology, applied in the field of resequencing microarray design, can solve problems such as increasing computational requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
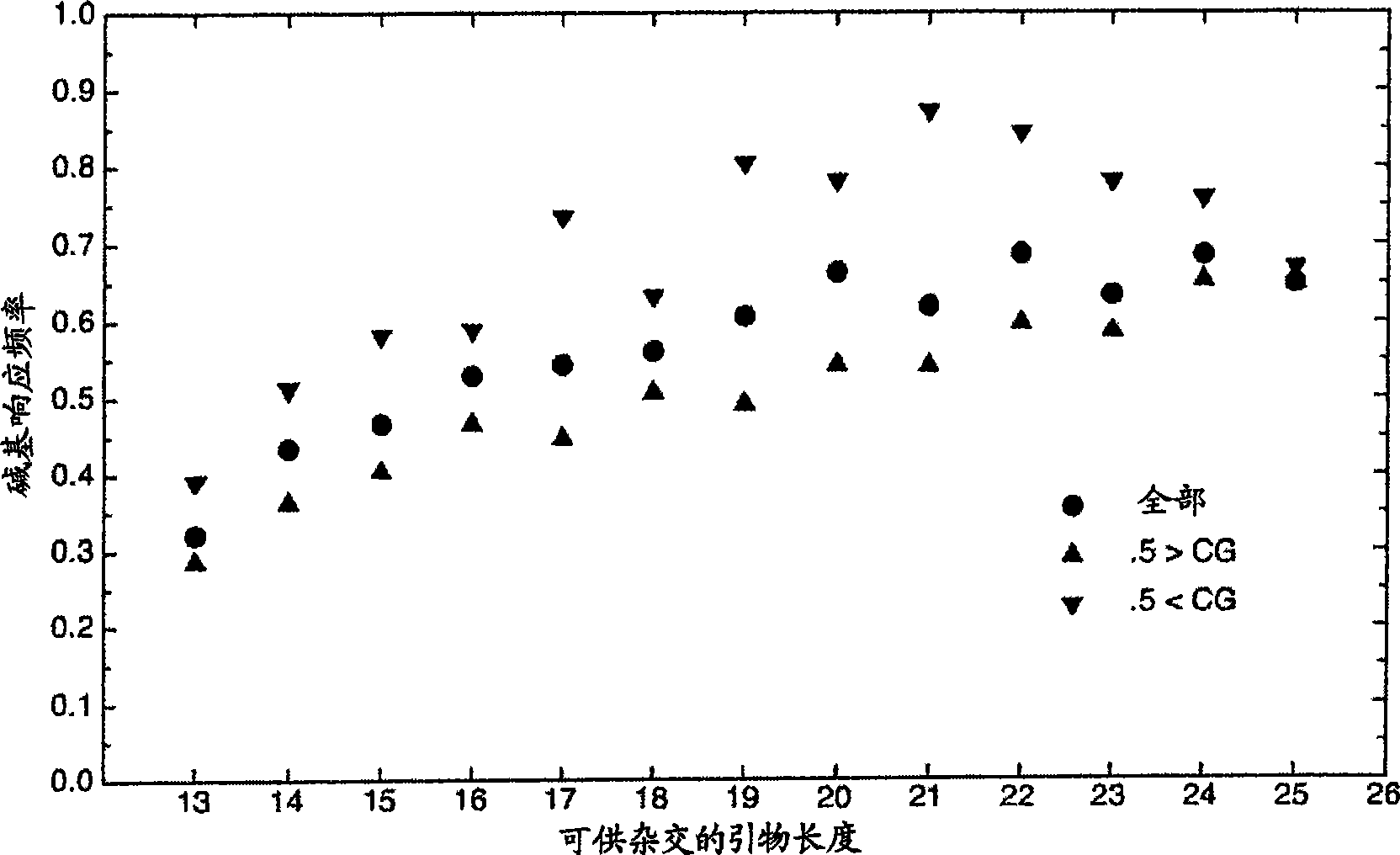
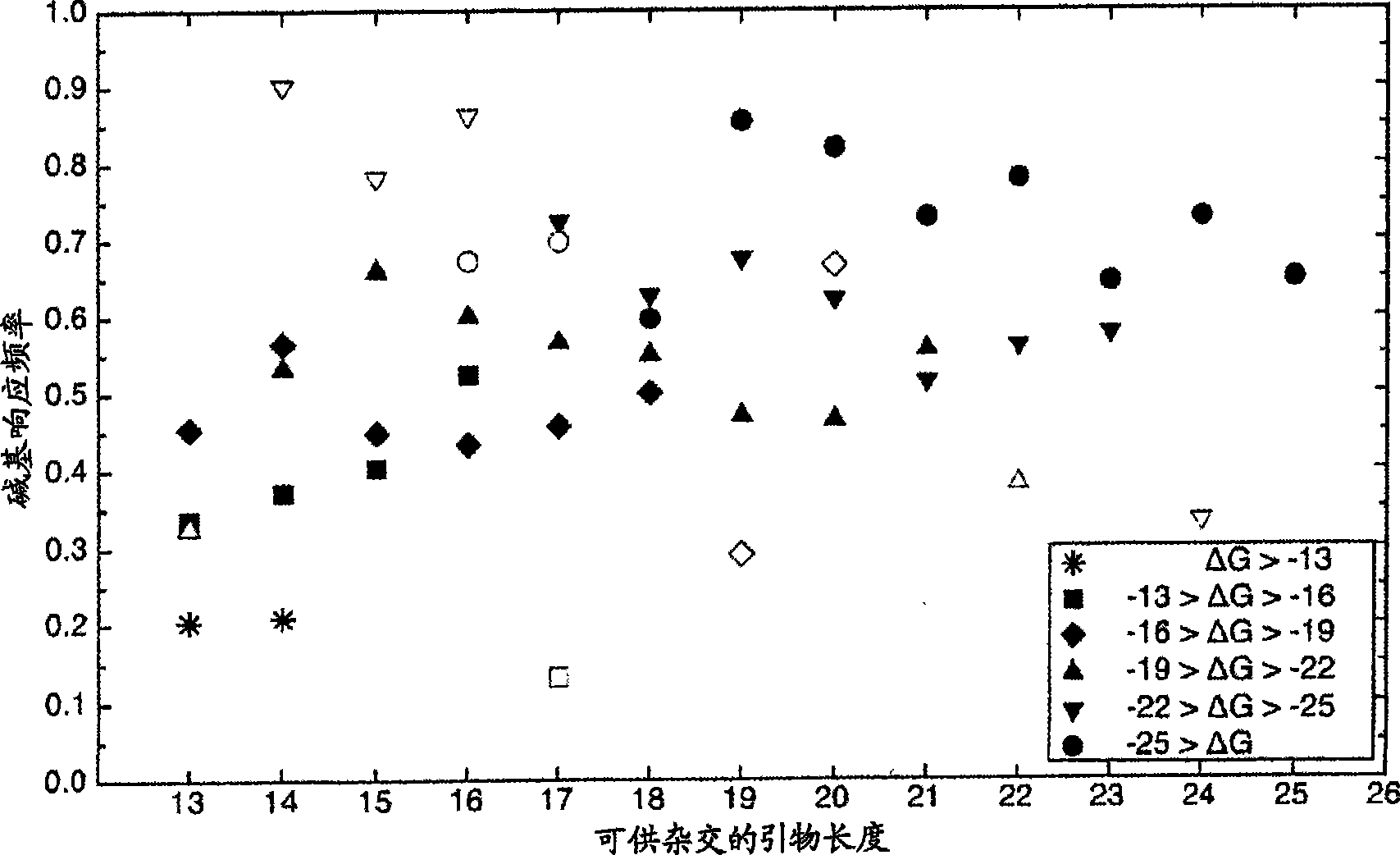
[0035] Example 1: Predicting Primer Interference - The first experimental use of the model algorithm was to understand the base calls that occurred in 42 microarray experiments with blank samples (no nucleic acid added) using the new primer set, where the The new primers try to minimize the interaction of the primers with the prototype sequence. Since the individual primers are still present, they are processed as a set of sample sequences and tested against each prototype sequence located on the chip using the model. The model accurately predicted base calls from primers that occurred in experiments that were still located on the prototype sequence. Additional binding to the central position of the prototype sequence was also observed and is in agreement with the experimental results. Primers designed to prototype sequences from closely related organisms elicit these base calls. For example, the prototype sequence of the adenovirus 4E1A gene allows 19 predicted bases out of...
example 2
[0036] Example 2: Model predictions on long sequences - After successfully demonstrating the accuracy of the model on shorter fragments, predictions on full prototype sequences were tested. The results of using routinely sequenced samples in the model are reported in Table 1, where the results are compared with experimental microarray results for 4 data sets; / H3N2 type California lineage, influenza virus B mountain county / 16 / 88 (Yamagata / 16 / 88) line and influenza virus B Victoria / 2 / 87 (Victoria / 2 / 87). The results report, for example, the general level of samples with great similarity to the Influenza A / H3N2 class Fujian samples, with an average base call rate of 85% for the assay, compared with an average of 97% for the model prediction. The average number of SNPs between the prototype and conventional sequences was 9.8 (1%). Although the model predicted that 9.2 SNPs would be resolved, only 6.3 SNPs were observed in the experiments. The model predicted 8.8 N calls for whic...
Embodiment 1
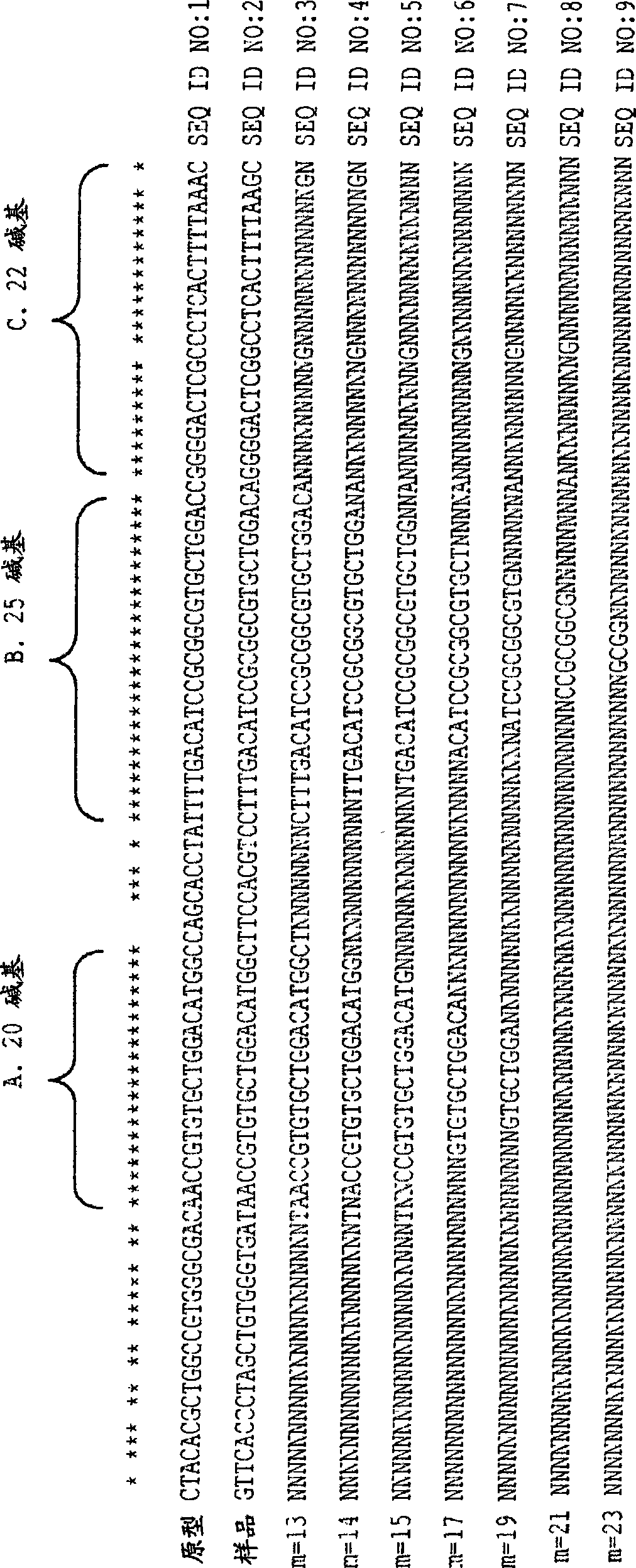
[0053]Hypothetical example with short sequences - The disclosed methods are illustrated below using artificial short sequences that would not correspond to any particular true species. There is a need to fabricate resequencing microarrays for detection of species A, B, C, D and E. As used herein, "species" may refer to taxonomic species as well as different types or strains of a single species and combinations thereof. Known nominal target 1 ( Figure 5 ) present in the genome of at least one of these species. Similarity sequence searches are performed using databases such as BLAST to generate target lists. A minimum similarity percentage (eg, 70%) can be used to filter out the results. If too many targets or targets from too many species (eg, genetically distant species) are reported, the minimum similarity percentage can be increased to reduce the size of the list. Additionally, the list can be manually checked to remove specific unfavorable targets.
[0054] Figure 5...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com