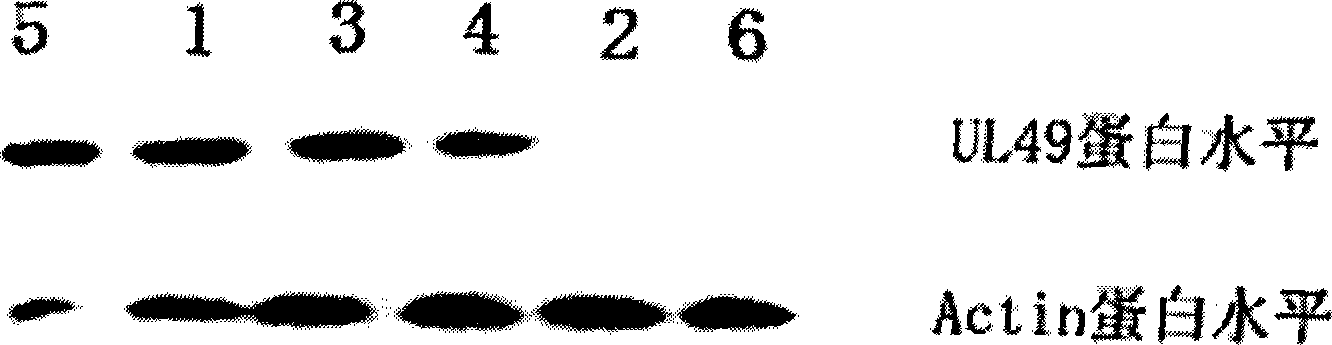External leader sequence for guiding RNase P ribozyme and use thereof in anti-HCMV medicament preparation
A technology of external guidance and nucleotide sequence, which is applied in the application field of preparing anti-HCMV drugs, can solve problems such as high cost, research and application obstacles, and achieve the effect of high drug resistance and cost reduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] The acquisition of embodiment 1 EGS
[0026] 1. Search for candidate EGS molecules in the mRNA sequence of the HCMV UL49 gene, and the search principles are as follows:
[0027] a. Search for UUCA or UUCG sequences in the target gene (UL49 gene) mRNA sequence;
[0028] b. The base anchoring the cleavage site within 20 to 25 bases upstream of the UUCR sequence is G;
[0029] c. The loop of the candidate EGS molecule must consist of 8-9 nucleotides;
[0030] d. The Steml of the candidate EGS molecule contains at least 1 G / C base pair, preferably between 3 and 4;
[0031] e. The candidate EGS sequence contains at least 7 to 8 base pairs;
[0032] f. The number of nucleotides between Stem1 and stem2 of the candidate EGS molecule and the target gene is preferably 5-6;
[0033] g. The Tm value of the candidate EGS molecule is around 50 degrees;
[0034] h. The binding position between the candidate EGS molecule and the target gene has less secondary structure.
[0035] ...
Embodiment 2
[0036] Example 2 Verification of the establishment of the EGS gene silencing efficiency platform
[0037] In this example, the UL49 gene was first cloned using a vector, and then a cell line capable of stably expressing UL49 was constructed. The vectors, cell lines, enzymes, and various reagents used in the example were all commercially available products. For example, pcDNA3.1(+ ) / myc plasmid, the cell line can choose Hela cell line, double enzyme digestion can use BamHI and XhoI, etc.
[0038] 1. Acquisition of UL49 gene
[0039] The UL49 gene can be amplified by PCR from the genome of the known sequence, for example, the UL49 gene can be amplified by PCR from the genome of the AD169 virus strain.
[0040] The total volume of the PCR reaction system is 100 μl: LA Taq enzyme 1 μl, reaction buffer 50 μl, AD169 genomic DNA template 5 μl, UL49 upstream primer (nucleotide sequence as shown in SEQ ID NO: 3) 5 μl, UL49 downstream primer (nucleotide sequence as shown in SEQ ID NO:...
Embodiment 3
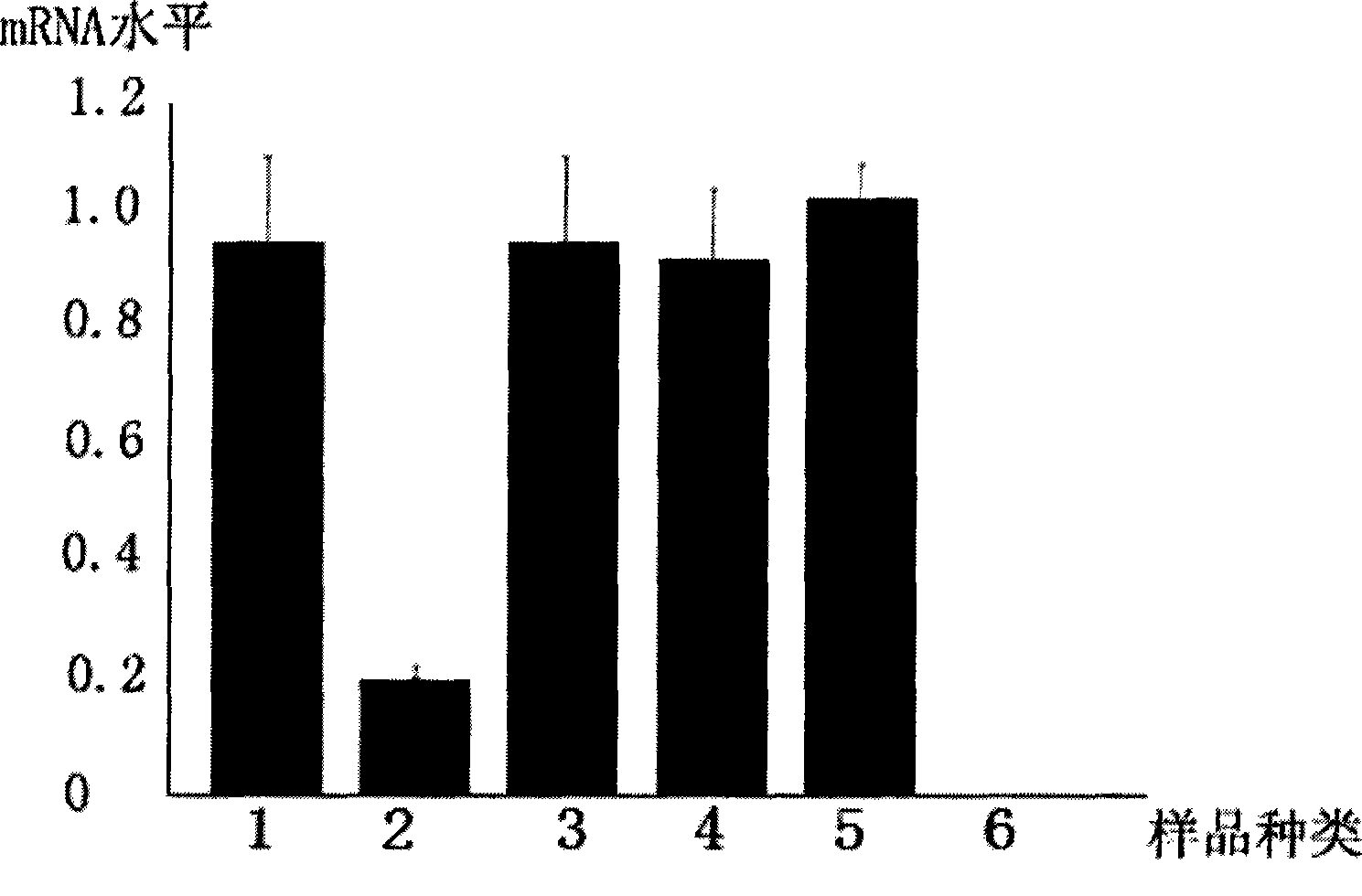
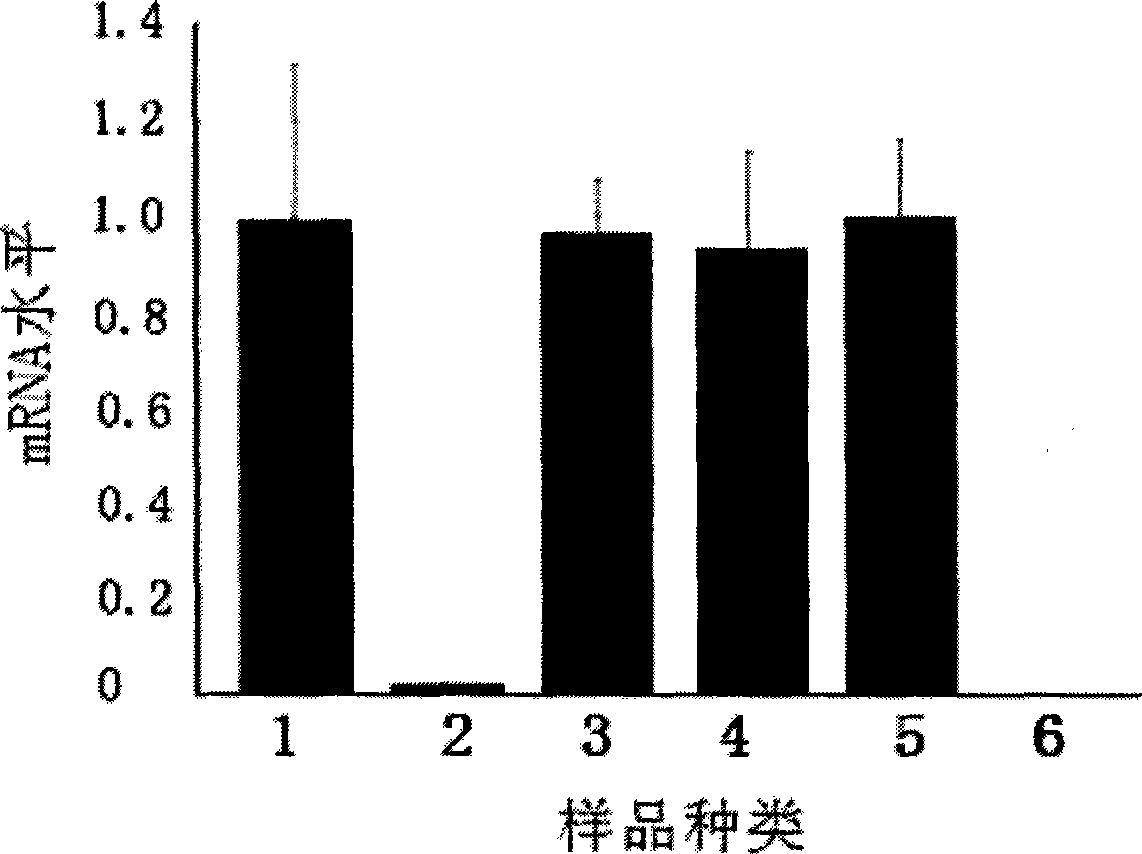
[0053] Example 3 EGS silences UL49 gene expression
[0054] The experimental group in this example is: the EGS prepared in Example 1 was transfected into the Hela cell line stably expressing the UL49 gene constructed in Example 2.
[0055] The control group of this embodiment is: (1) mutation control group: the mutant EGS prepared in Example 1 was transfected into the Hela cell line stably expressing the UL49 gene constructed in Example 2 according to the same transfection conditions as the experimental group; (2) Nonsense control group: the nonsense EGS prepared in Example 1 was transfected into the Hela cell line stably expressing the UL49 gene constructed in Example 2 according to the same transfection conditions as the experimental group; (3) Positive control group : the Hela cell line stably expressing UL49 constructed in Example 2, but not transfected with any EGS; (4) transfection reagent control group: refers to the stably expressing UL49 gene constructed in Example 2 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com