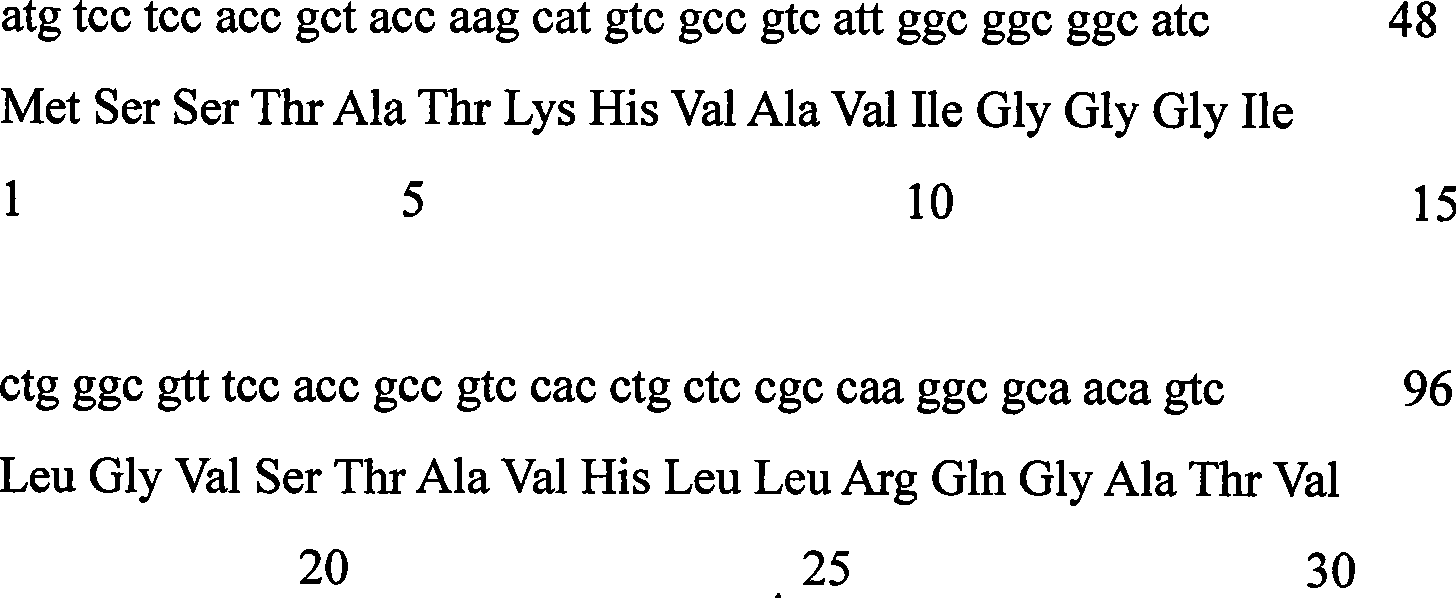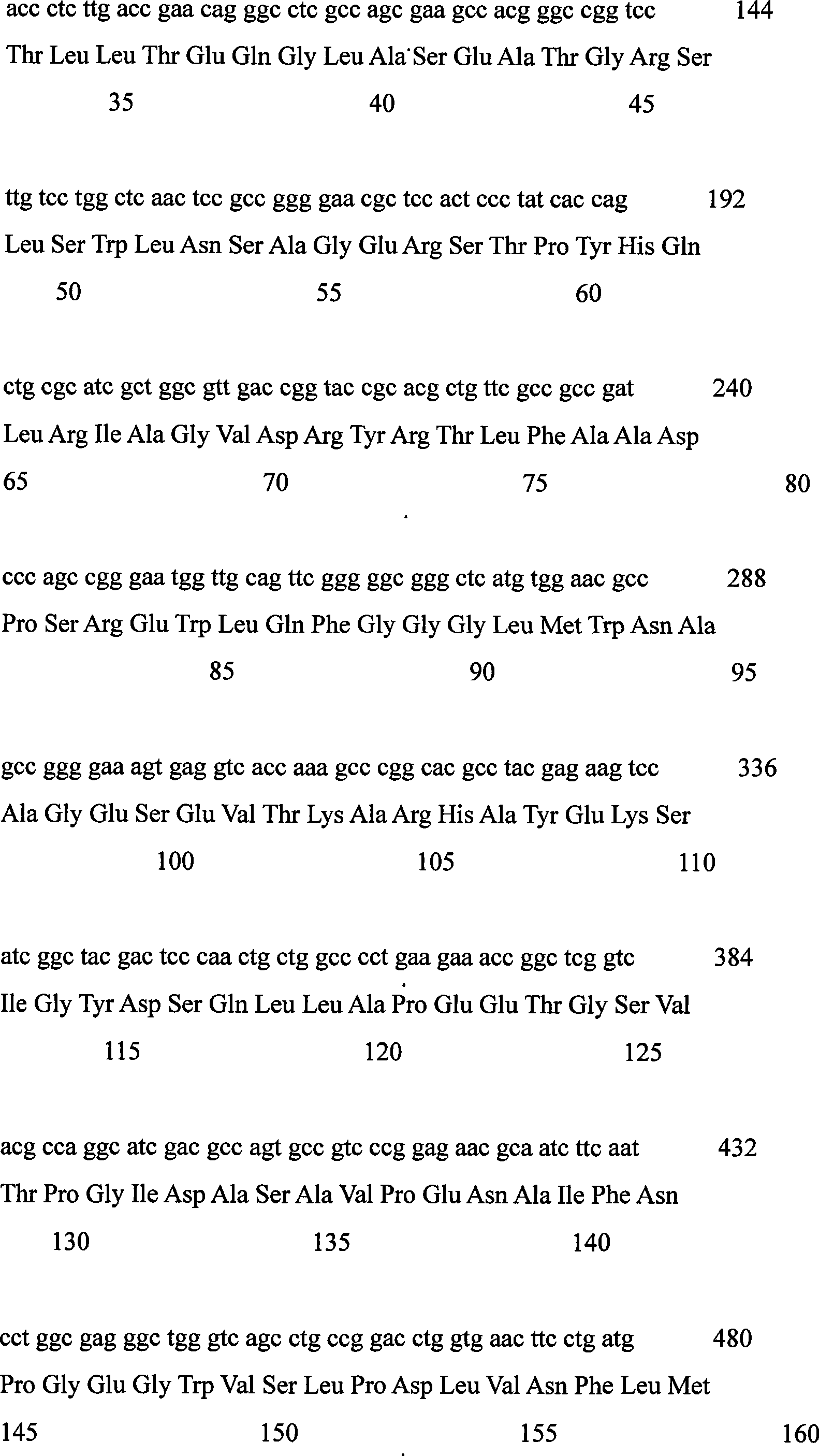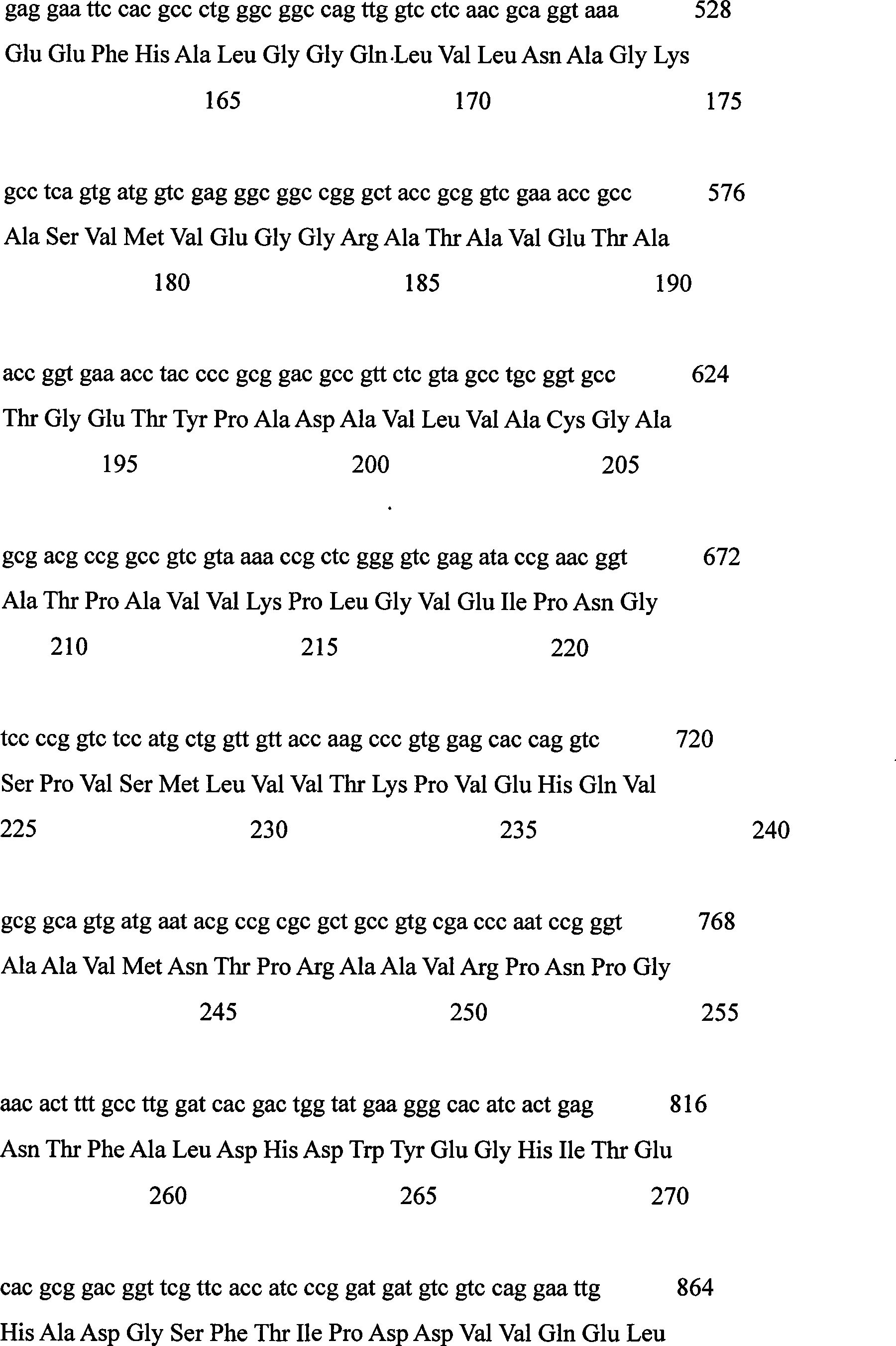Levulose valine oxidizing enzyme of high activity
A technology of valine oxidase and high activity, which is applied in the field of high activity fructose valine oxidase to achieve the effects of easy implementation, reduced enzyme amount and high activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Using the FVO gene coding sequence of Corynebacterium sp.2-4-1 as a template, error-prone PCR amplification was performed.
[0023] 1. Primer sequence:
[0024] Forward primer: 5'-TTGTTCGGATCCATGTCCTCCACCGCTAC-3'
[0025] Reverse primer: 5'-TTGTTCAAGCTTCTAGGAGAACCGGCCCG-3'
[0026] 2. Error-prone PCR reaction system and reaction conditions:
[0027] PCR reaction system:
[0028] 100μL system contains:
[0029] 10mM Tris-HCl pH8.30, 50mM KCl, 6.5mM MgCl 2 ,
[0030] 0.15mM MnCl 2 , 0.2mM dGTP / dATP, 0.8mM dTTP / dCTP,
[0031] 2.2μg SSB Protein, 0.5μM forward / reverse primer, 10ng template DNA
[0032] 2.5 units of Taq DNA polymerase.
[0033] PCR reaction conditions:
[0034] 94°C for 5min; 94°C for 30sec, 55°C for 30sec, 72°C for 2min, 35 cycles; 72°C for 10min; 4°C forever.
[0035] 3. Take 3 μL of the error-prone PCR product and electrophoresis on 1% agarose gel. It can be seen that there is a product band of about 1 kb. The error-prone PCR product was purifi...
Embodiment 2
[0037] Initial screening of mutant strains:
[0038] Extract the plasmid DNA in the mutant library, digest it with BamH I and Hind III, and recover the target fragment of about 1kb. The vector pGEX-4T-1m is also digested with BamH I and Hind III, recover, and connect the two at 4°C Overnight, transformed into BL-21 strain, spread on LB containing IPTG, fructose valine and N-(carboxymethylaminocarbonyl)-4,4-bis(methylamino)-benzidine [DA-64] Plates, after 12-24 hours of incubation at 37 ° C, obvious discoloration can be seen. The clones with fast discoloration and large discoloration circle were picked for liquid medium culture, and a total of 48 clones were picked.
Embodiment 3
[0040] Quinone method detection of mutant enzyme activity:
[0041] 1. After inducing expression for 4 hours, collect the bacterial liquid, measure and record the OD600 value of the cell culture.
[0042] 2. Prepare bacterial lysate for detecting enzyme activity. In 374 μL B-PER TM Cells were resuspended in bacterial protein extraction reagent (Pierce product78248), and then 50uL of protease and phosphorylase inhibitors and bacterial cell extract (Sigma product P8465), and 1uL of 34mg / mL chloramphenicol (prepared with methanol) were added. Vortex quickly for one minute. Place on ice for 5 min. Centrifuge for 1 min and place on ice.
[0043] 3. Determination of protein content by Bradford method.
[0044] 4. Take 50 μL of the above bacterial lysate and add it to the quinone method reaction mixture, incubate at 37°C for 1-3min, and measure the absorbance at 555nm. The composition of the reaction mixture is: 100mM potassium phosphate buffer (pH8.0), 1purpurogallin unit / ml p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com