Human tumour specific Ki67 gene promotor
A tumor-specific, promoter technology, applied in the field of medical genetic engineering, can solve the problems of unclear biological function and little research, and achieve the effect of good application value, high tumor specificity, and high promoter activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Embodiment 1 Contains the construction of the eukaryotic expression plasmid of human Ki67 gene promoter
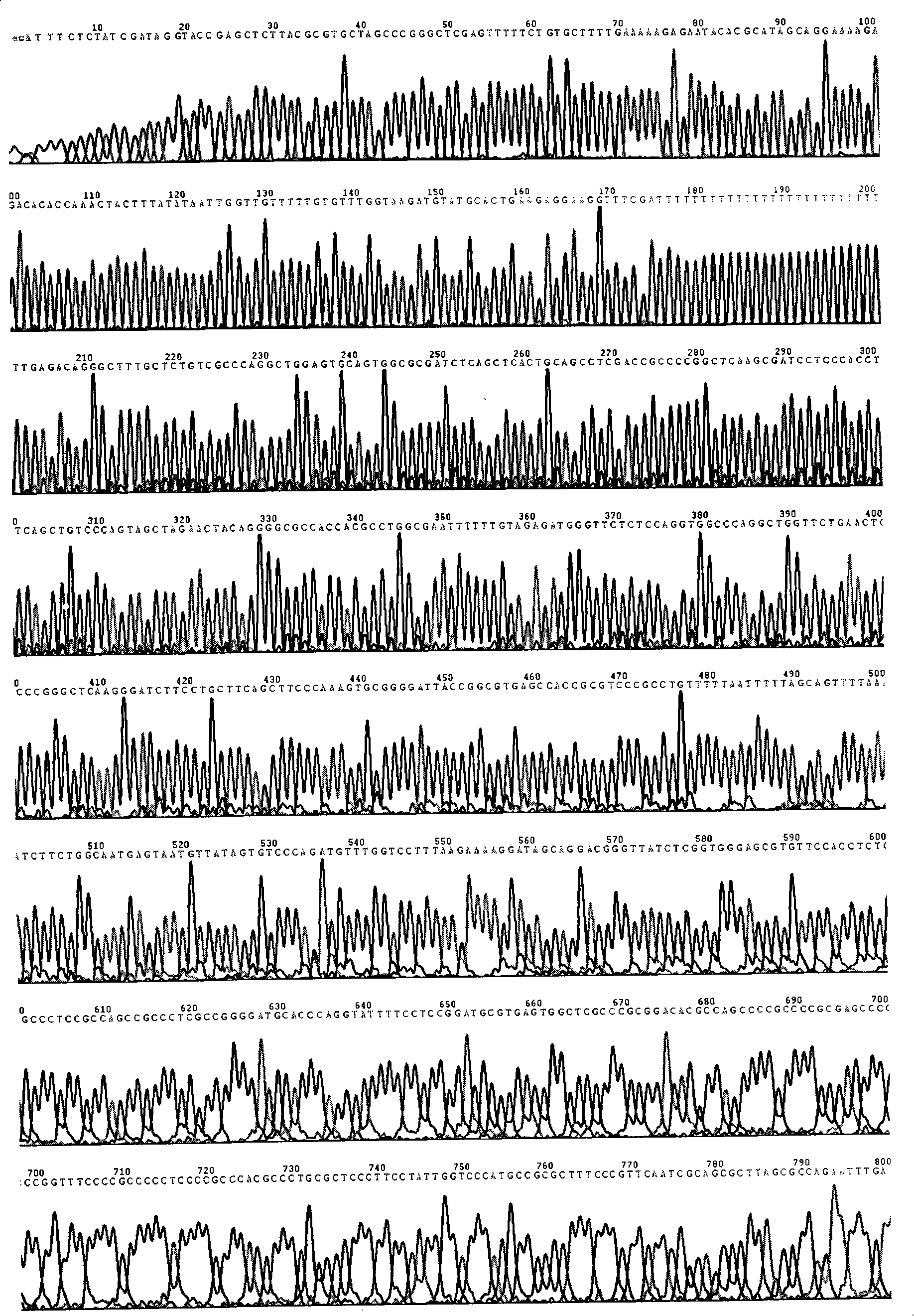
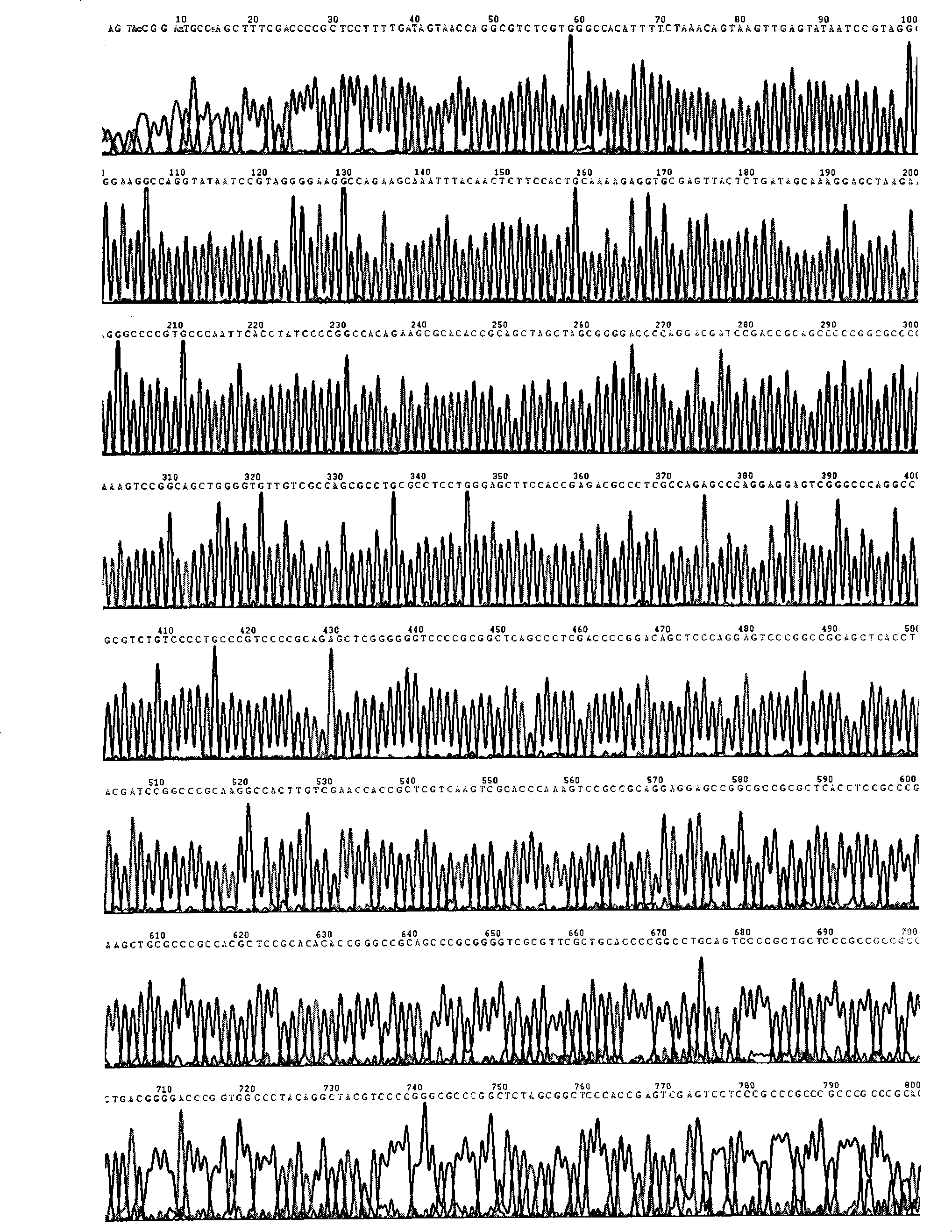
[0027] 1. Cloning and sequence identification of the 5' flanking sequence (-820~+771) of the human Ki67 gene
[0028] The 5' flanking sequence of the transcription start point of Ki67 gene was searched in Genbank, and a pair of primers were designed and synthesized to amplify the 1591 fragment upstream of the start codon of Ki67 gene. Design upstream primers in the -820~-805 region, and introduce the Xho I restriction site, design downstream primers in the +758~+771 region, and introduce the Hind III restriction site. The upstream primer is named KU800, the sequence is: 5′-ttgt ctcgagtttttctgtgcttttg-3′, the downstream primer is named KD1, and the sequence is: 5′-tgtg aagctttcgaccccgctcct-3′.
[0029] Using this pair of primers, using the genomic DNA of human kidney cancer Ketr-3 cells as a template, a DNA fragment of about 1.6kb was amplified by PCR, and the targe...
Embodiment 2
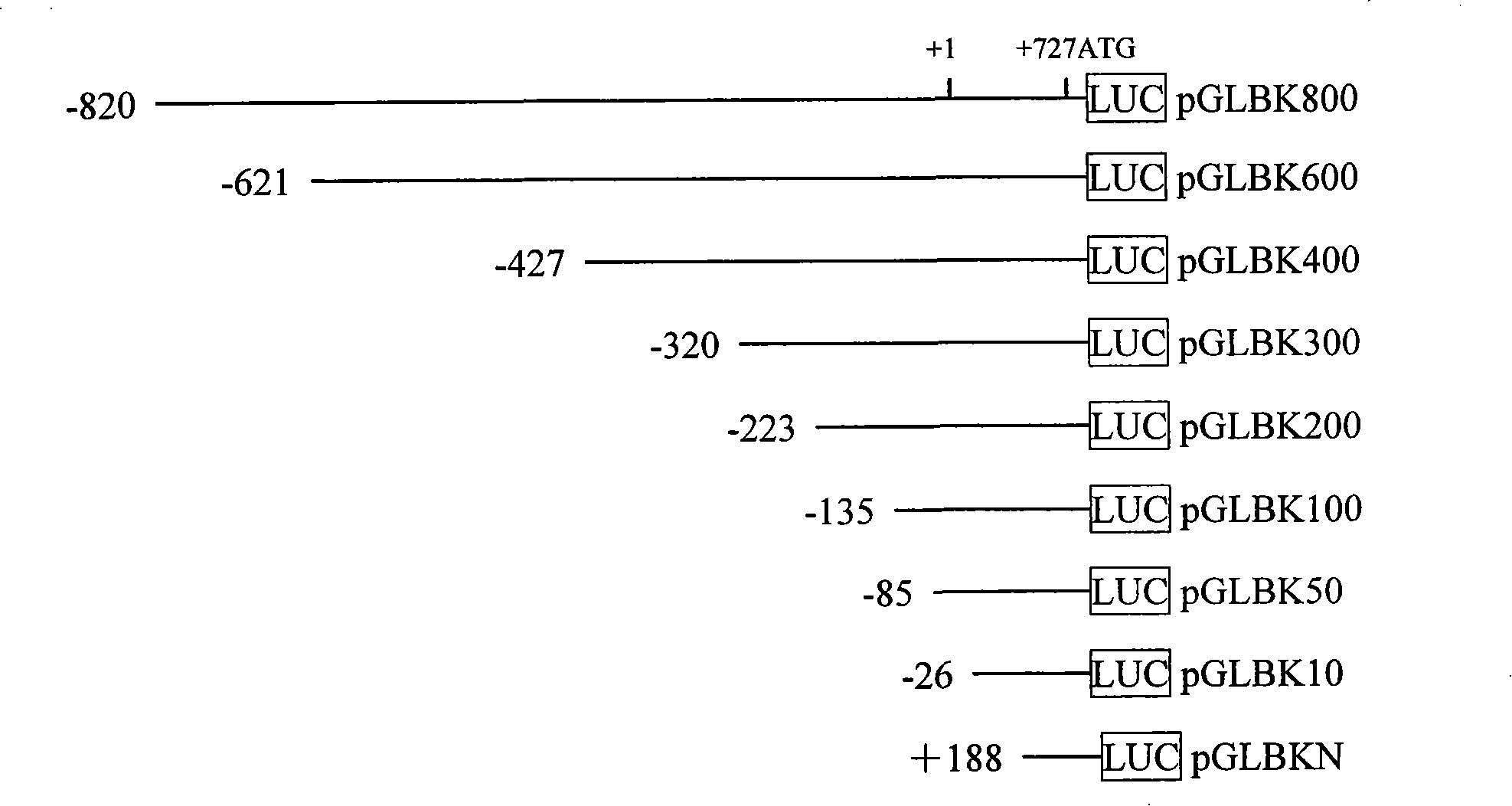
[0040] Example 2 Cell culture, plasmid transfection and human Ki67 gene promoter activity analysis
[0041] After each experimental plasmid (eukaryotic expression plasmid of the Ki67 promoter fragment) was successfully constructed, the plasmid was extracted with the PureYieldPlasmid Midiprep System kit, and the positive control plasmid pGL3-Control (containing SV40 promoter / enhancer) and the negative control were extracted at the same time. Plasmid pGL3-Basic (without SV40 promoter / enhancer) and internal reference plasmid pRL-TK (expressing renilla luciferase under the control of HSV-TK promoter). The experimental plasmid and control plasmid were diluted to 100ng / μl, and the internal reference plasmid pRL-TK was diluted to 20ng / μl.
[0042] In the present invention, each experimental plasmid and control plasmid were respectively transfected into human cervical cancer cells (Hela), bladder cancer cells (BIU-87), renal cancer cells (Ketr-3), lung cancer cells (A549) and human um...
Embodiment 3
[0057] Embodiment 3 Ki67-promoter, hTERT promoter, Survivin promoter activity comparison
[0058] In order to further verify the magnitude of Ki67-promoter transcriptional activity, we compared it with the two tumor-specific promoters hTERT and Survivin promoters that have been studied more. Experimental method and steps are the same as in Example 2. The results showed that in all tumor cells, the activity of Ki67-promoter was stronger than that of hTERT and Survivin promoters, and in normal cells, all three promoters were inactive (see Table 6, Figure 12 , 13 ). Ki67-promoter showed better promoter activity.
[0059] Table 6 The activity comparison of Ki67 core promoter, hTERT promoter and Survivin promoter in each cell
[0060]
[0061] b Ratio of relative luciferase activity to pGL3-Control plasmid
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com