Method for rapidly detecting impatiens necrotic spot virus from oncidium
A detection method and technology of oncidium, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of small transmission medium, harm to orchid industry, fast transmission and reproduction speed, etc. The effect of improved sex and specificity, high sensitivity and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] ——RT-PCR detection
[0022] Samples were taken from the symptomatic part of the susceptible plant, the leaf tip far away from the symptom, and the healthy plant, and the total plant RNA was extracted. After synthesizing cDNA, conventional PCR amplification was performed using ZI2F and ZI2R as specific primers. PCR reaction system 25 μL: 2 μL of the above reverse transcription product, 3 μL of 10×PCR Buffer, Mg 2+ (25mM) 1.2μL, dNTP Mixture (2.5mM each) 0.4μL, ZI2F 0.4μL, ZI2R 0.4μL, Taq enzyme (5U / μL) 0.2μL and sterile water 17.4μL. Reaction program: 94°C for 5min; 30 cycles of 94°C for 30s, 45°C for 30s, and 72°C for 40s; 72°C for 8min.
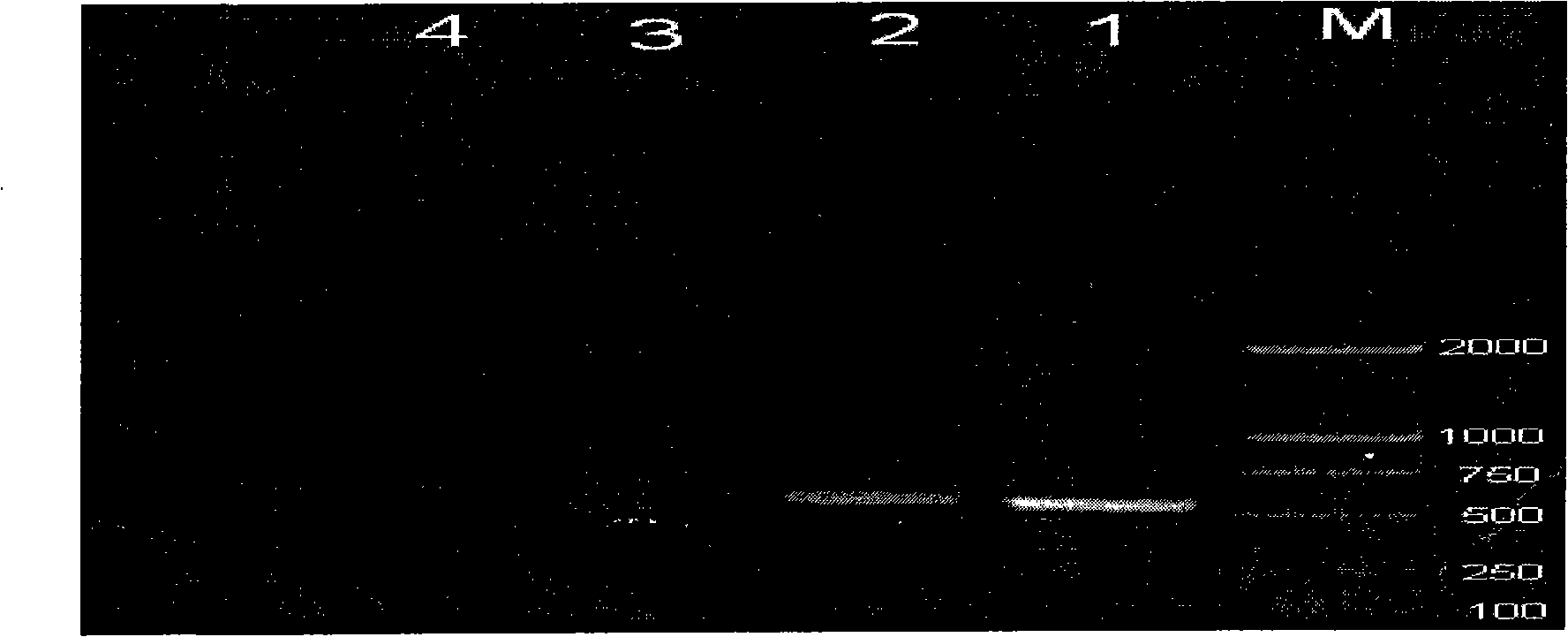
[0023] RT-PCR amplification products were detected by 1% agarose gel electrophoresis, and the detection results were as follows: figure 2 , after PCR amplification on the leaves showing symptoms, a target band consistent with the expected fragment size can be seen at 500bp (see figure 2 , lanes 1, 2). Target bands with the expec...
Embodiment 2
[0026] ——Nested PCR detection
[0027] The first round of PCR amplification was performed with ZI1F and ZI1R as outer primers, and the second round of amplification was performed with ZI1F and ZI3R, ZI3F and ZI1R, ZI3F and ZI3R as inner primers respectively. Template 2μL, 10×PCR Buffer 3μL, Mg 2+ (25mM) 1.2μL, dNTP Mixture (2.5mM each) 0.4μL, upstream primer 0.4μL, downstream primer 0.4μL, Taq enzyme (5U / μL) 0.2μL and sterilized water 17.4μL. Reaction program: 94°C for 5min; 30 cycles of 94°C for 30s, 52°C for 30s, 72°C for 40s; 72°C for 8min.
[0028] At the same time, the cDNA was multiplied by 10 times, 10 -2 times, 10 -3 times, 10 -4 times, 10 -5 After two-fold dilution, primers ZI1F and ZI1R were used for the first round of amplification, and then ZI3F and ZI3R were used as primers for the second round of amplification to verify the sensitivity of nested PCR.
[0029] 1% agarose gel electrophoresis detection results showed that ZI1F and ZI3R amplified fragments 850b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com