Multiple real time fluorescence quantifying PCR method for detecting porcine circovirus, porcine parvovirus, porcine pseudorabies virus and classical swine fever virus
A porcine pseudorabies virus, real-time fluorescence quantitative technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as dealing with cross-contamination, harm, etc., and achieve the effect of good application stability and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 Standard plasmid preparation
[0040] 1 Preparation of target fragments The nucleic acids of PRV, PPV, PCV-2, and HCV were extracted according to conventional methods, and the nucleic acids were stored at -20°C for later use. To design primers, use OLIGO6.24, compare the nucleic acid sequences in GENEBANK, and design primers (Table 1). The extracted PRV, PPV, PCV-2, and HCV viral nucleic acids were used as templates, and the primers in Table 1 were used for PCR amplification. The length of the amplified products was expected to be 265bp, 764bp, 792bp and 303bp. The amplified products obtained were subjected to agarose gel electrophoresis, and the results were shown in figure 1 .
[0041] Table 1 Sequences of primer pairs for amplifying target genes
[0042]
[0043] 2 Preparation of standard plasmids Recover the target fragments according to the Plasmid Miniprep Kit operation method, connect the recovered products to the pMD18-T vector, transform Escher...
Embodiment 2
[0045] Example 2 Establishment of SyMuReTi-PCR detection method for 4 kinds of viruses
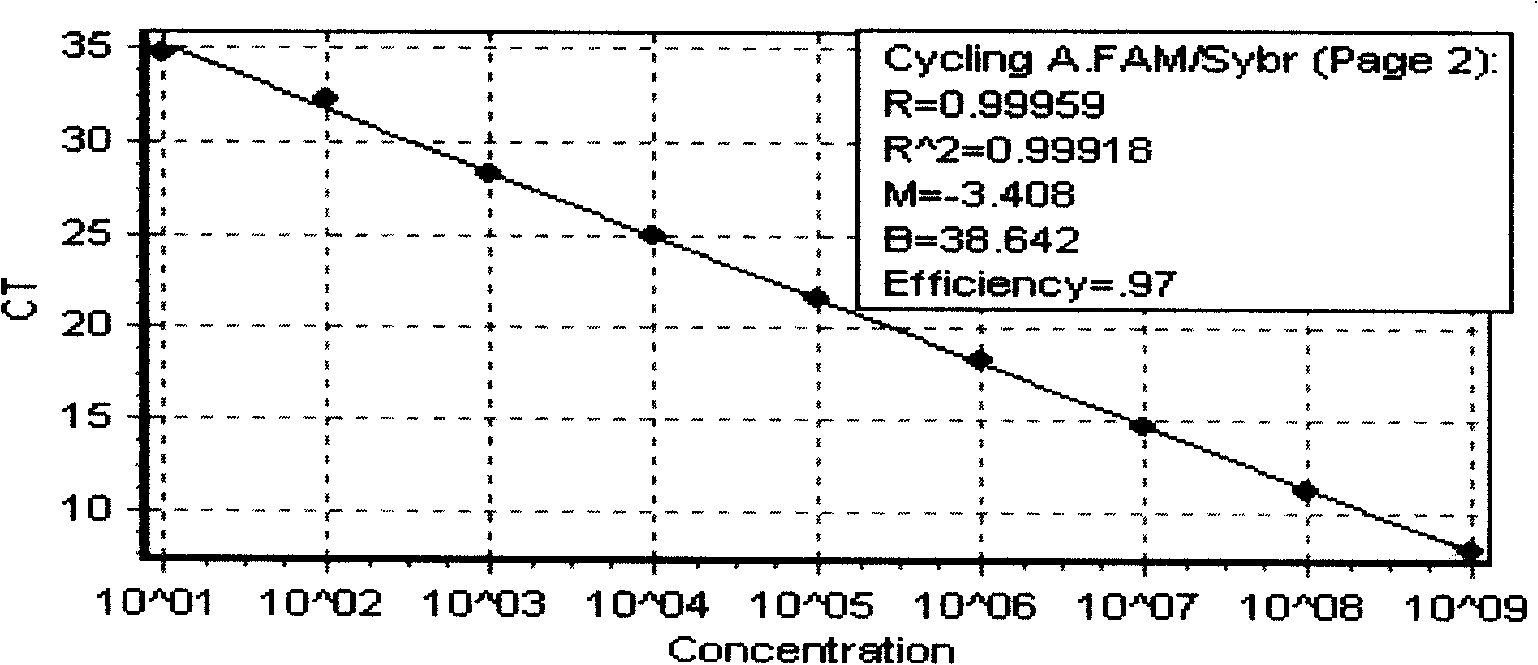
[0046] 1.1 Dilution of the standard plasmids of four kinds of viruses The quantitative standard plasmid is 4.97×10 using Eppendorf Biophotometer 12 Copies / μl, 3.44×10 12 Copies / μl, 8.18×10 12 Copies / μl, 7.98×10 12 Copies / μl, use EASY Dilution to dilute the standard plasmid at 1.0×10 8 ~1.0×10 1 Copies / μl 10-fold dilution, stored at -20°C.
[0047] 1.2 Detection primer design The target gene cloned in Example 1 was sequenced, and four pairs of specific primers were designed using OLIGO6.24 according to the determined sequence (Table 2).
[0048] Table 2 Sequences of primer pairs for amplifying target genes
[0049]
[0050] 1.3 Add SyMuReTi-PCR to an Axygen Scientific Quality centrifuge tube (200 μl) in sequence: 9.5 μl of sterilized deionized water, 1 μl of dimethyl sulfoxide, 0.5 μl of each of the 4 pairs of primers in Table 2, and add different dilutions to different centrifuge ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com