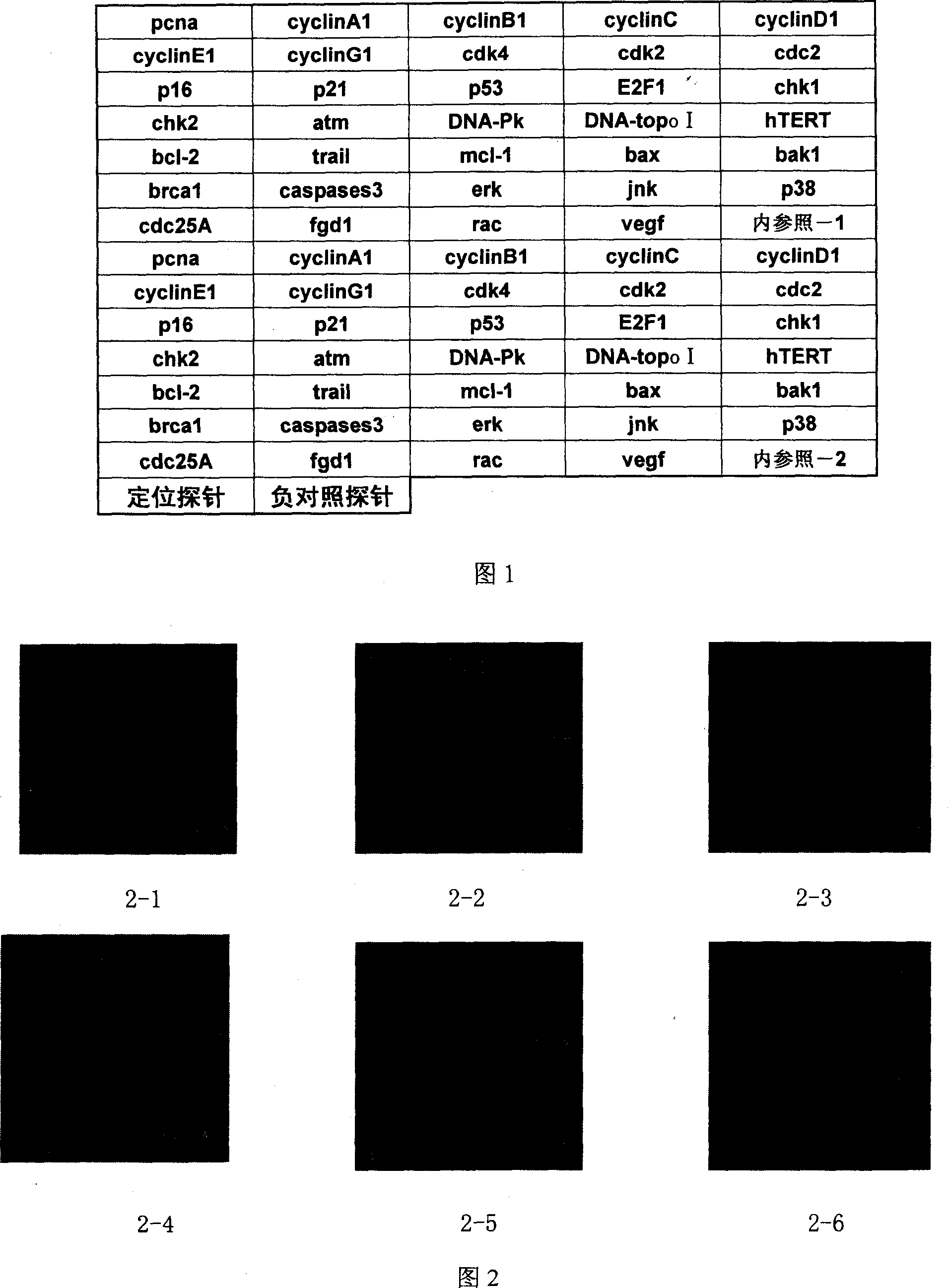
Biological chip for detecting control of related gene expression by pharmaceutical active component
A gene expression and biochip technology, which is applied in the field of biochips to detect the regulation of drug active ingredients on the expression of related genes, can solve the problems of numerous genes, unsuitable applications, and high costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] drug treated cells
[0082] 1. Cell culture
[0083] K562 cells were cultured in RPMI1640 (pH 7.0) medium containing 10% fetal bovine serum and 80 U / L gentamicin at 37°C and 5% CO 2 Growth was incubated in a constant temperature incubator.
[0084] 2. Treatment of cells with antineoplastic drug artemisinin
[0085] Weigh a certain amount of penicillin and dissolve it in 500 μl DMSO, and heat to help dissolve it. Then diluted with serum-free medium to a concentration of 256×10 -5 mol / L. Before use, dilute to the required concentration with serum-free medium, respectively 1×10 -5 mol / L, 4×10 -5 mol / L, 16×10 -5 mol / L, 64×10 -5 mol / L, 256×10 -5 mol / L. Routinely cultured cells, adjusting the number of cells to 1×10 5 Each / mL, add 0.5ml of the above-mentioned drugs of each concentration. to a final concentration of 1 x 10 -6 mol / L, 4×10 -6 mol / L, 16×10 -6 mol / L, 64×10 -6 mol / L, 256×10 -6 mol / L. The cells were then placed at 37°C, 5% CO 2 Incubate in the inc...
Embodiment 2
[0087] TRIzol reagent for RNA extraction
[0088] Collect the cells into a centrifuge tube after 24 hours of drug action, discard the culture medium, invert to air dry, add 1ml TRIzol reagent, pipette repeatedly, transfer to a 1.5ml Eppendorf tube, and place at 15-30°C for 5 minutes. Add 0.2ml of chloroform (trichloromethane), vibrate vigorously for 15 seconds; then continue to stand at 15-30°C for 2-3 minutes; centrifuge at 12000×g for 15 minutes at 2-8°C. After centrifugation, take the upper colorless RNA-containing aqueous phase (about 360ul) to a new 1.5ml Eppendorf tube. Do not suck the solution below the water phase, so as not to contaminate DNA and protein. Add 0.5ml of isopropanol, let stand at 15-30°C for 10 minutes, then centrifuge at 12000×g for 10 minutes at 2-8°C. Discard the supernatant, add at least 1ml of pre-cooled 75% ethanol to wash the RNA pellet once. Shake vigorously to mix the sample and centrifuge at 2-8°C at least 7500×g for 5 minutes. Discard the ...
Embodiment 3
[0090] cDNA generation - reverse transcription direct labeling experiments
[0091]Preparation system 1: 10-40 μg total RNA, ≤11 μl; Anchored Oligo (dT) 20 Primer (2.5 μg / μl), 2 μl, RNase-Free water to 13 μl, mix well, briefly centrifuge for 5 seconds, heat at 70°C for 10 minutes, then quickly Place on ice and incubate for at least 1 min. Configuration System 2: System 1, 13μl; 5X First-Strand buffer, 6μl; (0.1M) DTT3μl, dNTP Mix for labeled, 2μl; RNaseOUT TM (40U / μl), 1μl; Labeled dCTP (1mM) 3μl; SuperScript TM III RT (400U / μl), 2μl; total volume 30μl, mix well, briefly centrifuge for 5 seconds, incubate at 46°C for 3 hours, and protect from light during the incubation period. After the incubation, add 15ul of 0.1M NaOH, mix well, and heat at 70°C for 30 minutes. Add 15ul 0.1M HCl and mix well.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com