Clear Cell Renal Cell Carcinoma Biomarkers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
and Methods
[0082]Fresh-frozen normal-tumour tissues were obtained from nephrectomy cases under approvals from institutional research ethics review committees and patient consent. Normal tissues were harvested from sites distant from the tumour. Table 1 refers to detailed patient information of this study.
TABLE 1Patient informationAge at theFuhrmantime ofNuclearIDdiagnosisRaceSexHisto TypeGrade7457585980ChineseFemaleClear cell carcinomaGrade II1762195348ChineseMaleClear cell carcinomaGrade II5739866756ChineseMaleClear cell carcinomaGrade III7052883552ChineseMaleClear cell carcinomaGrade III7797208350ChineseMaleClear cell carcinomaGrade IV7541692349Un-MaleClear cell carcinomaGrade IIknown2043171365IndianMaleClear cell carcinomaGrade IVwith pappilaryfeatures4091143256ChineseFemaleClear cell carcinomaGrade II12364284Un-Un-Un-Clear cell carcinomaGrade IIIknownknownknown8604910251ChineseMaleClear cell carcinomaGrade II
Cell Lines
[0083]Commercial cell lines (786-O, A-498,...
example 2
atory Landscapes in Clear Cell Renal Cell Carcinoma Tumours are Aberrant
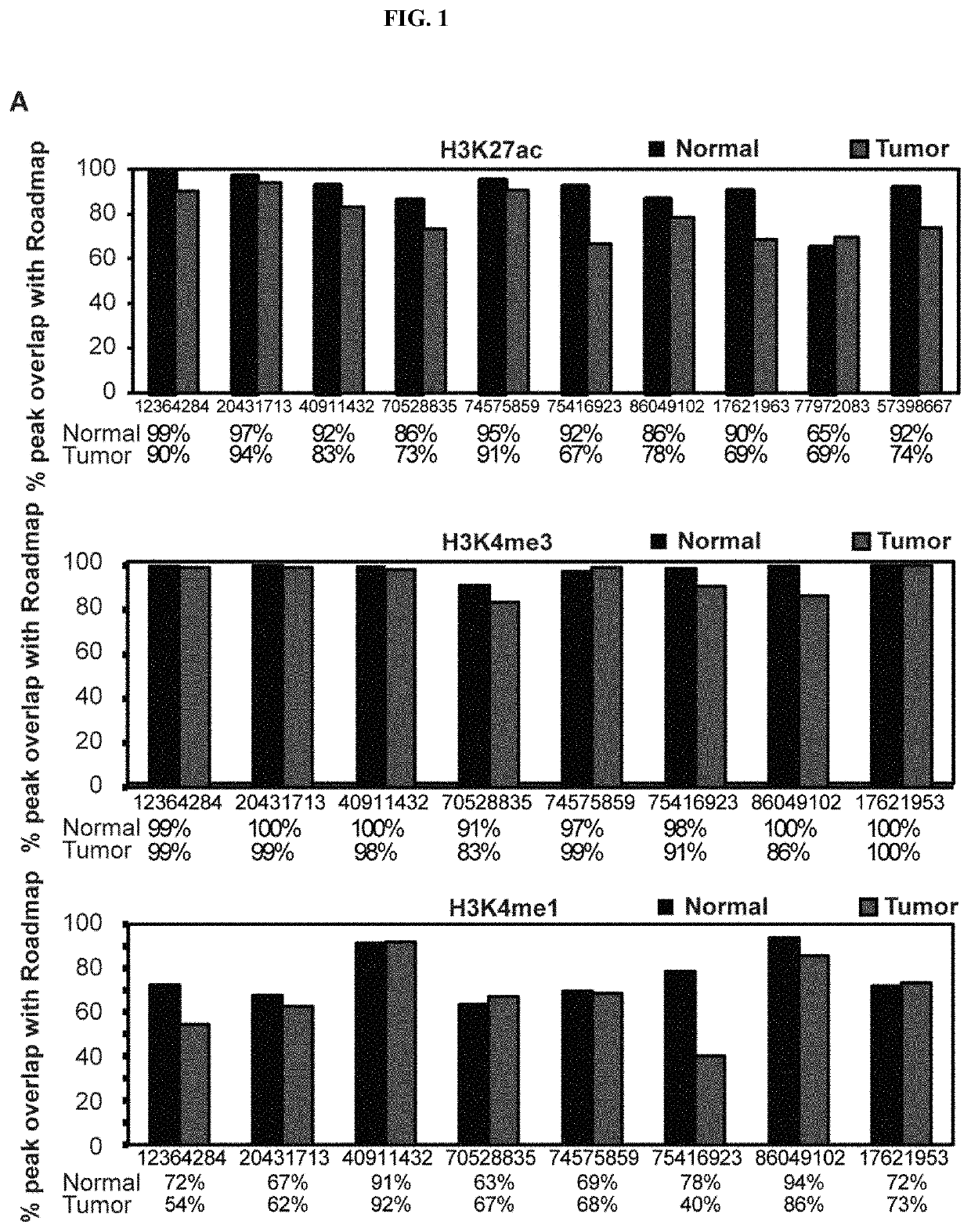
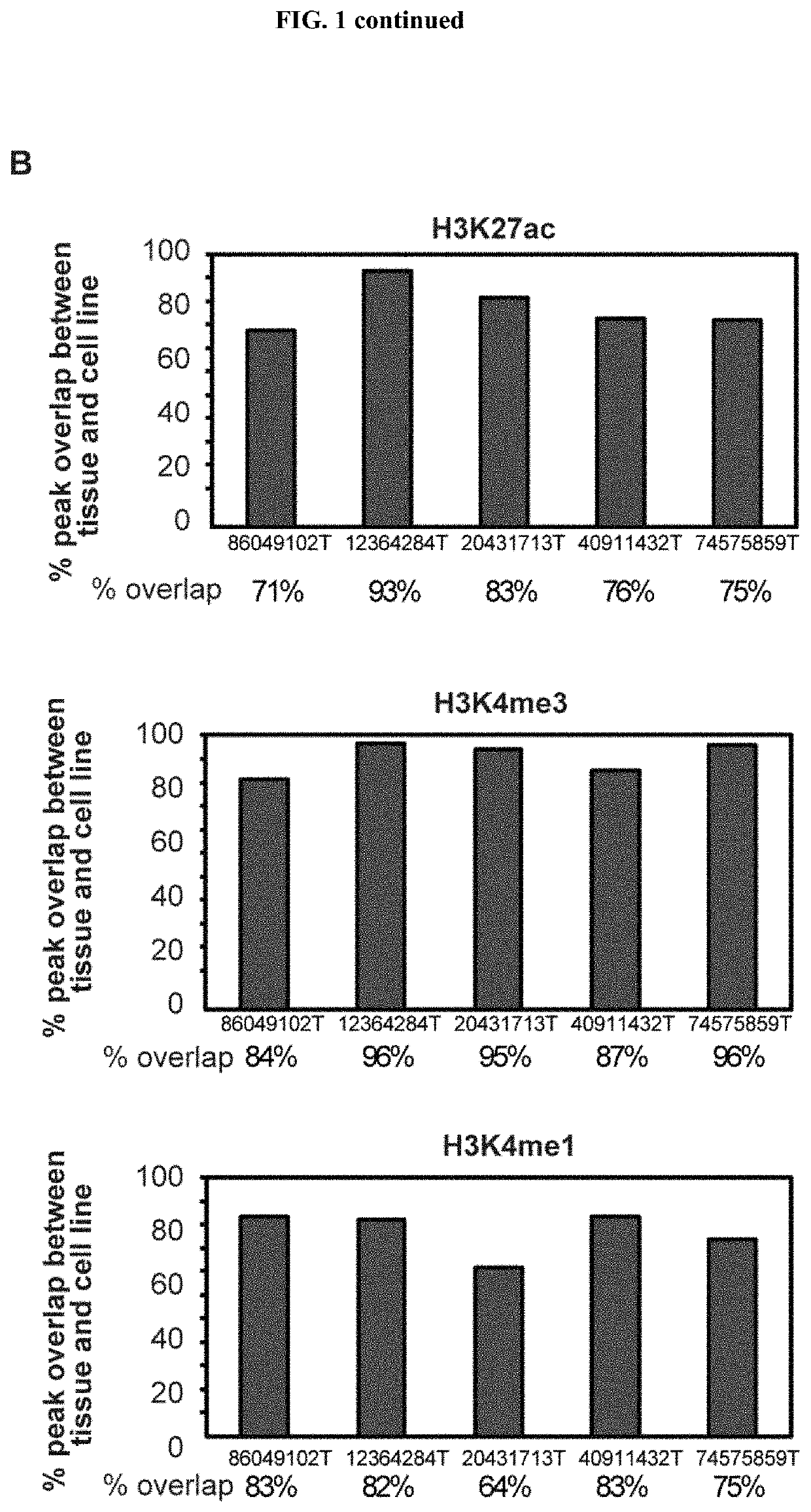
[0123]To explore whether clear cell renal cell carcinoma tumours display alterations in their cis-regulatory landscapes in vivo, histone chromatin immunoprecipitation sequencing (ChIP-seq) profiles (3 marks: H3K27ac, H3K4me3, and H3K4me1) were generated in 10 primary tumour / normal pairs, 5 patient-matched tumour-derived cell lines, 2 commercially available clear cell renal cell carcinoma lines (786-O and A-498), and 2 normal kidney cell lines (HK2 and PCS-400. Table 1 in example 1 shows patient clinical information. Of the original 87 samples, 79 samples passed pre-sequencing quality-control filters and were subjected to ChIP-seq processing and downstream analysis. In total, 2,363,904,778 uniquely mapped reads were generated. On average, 89% of H3K27ac peaks, 98% of H3K4me3 peaks, and 76% of H3K4me1 peaks obtained in our normal kidney tissues overlapped with peaks from adult kidney tissues in the Epigenomics Roa...
example 3
ecific Enhancers are Associated with Hallmarks of Clear Cell Renal Cell Carcinoma
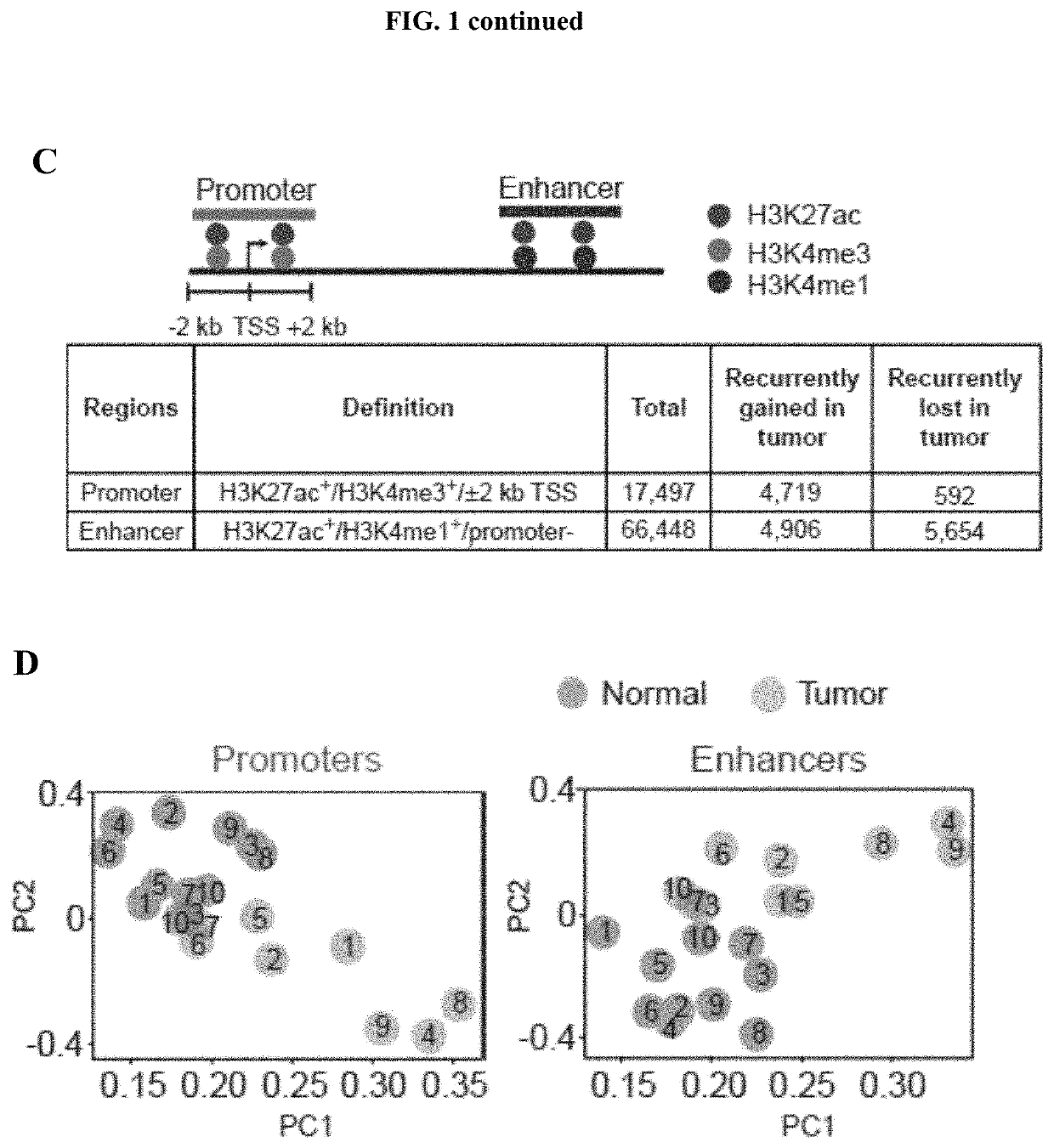
[0127]To identify genes modulated by the tumour-specific regulatory elements, enhancers were assigned using three approaches. The first approach utilized predefined linear proximity rules involving a set of highly confident genes (GREAT algorithm). MSigDB pathway analysis using GREAT-assigned genes revealed that gained enhancers exhibit a highly significant renal cell carcinoma-specific signature compared with gained promoters (enhancer q-value=3.2×10−26; promoter q-value=1.5×10−1, binomial FDR; FIG. 2A). Although gained promoters were involved in general cancer processes (for example, cell cycle, transcription, and RNA metabolism) for a complete list of promoter pathways), gained enhancers were enriched in disease-specific features of clear cell renal cell carcinoma, including HIF1α network activity, proangiogenic pathways (platelet activation and PDGFRβ signaling), and SLC-mediated transmembrane trans...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com