Purification of immunoglobulins using affinity chromatography and peptide ligands
a technology of affinity chromatography and immunoglobulin, which is applied in the field of purification of immunoglobulins using affinity chromatography and peptide ligands, can solve the problems of contaminating products, low stability, and high cos
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Peptide Synthesis
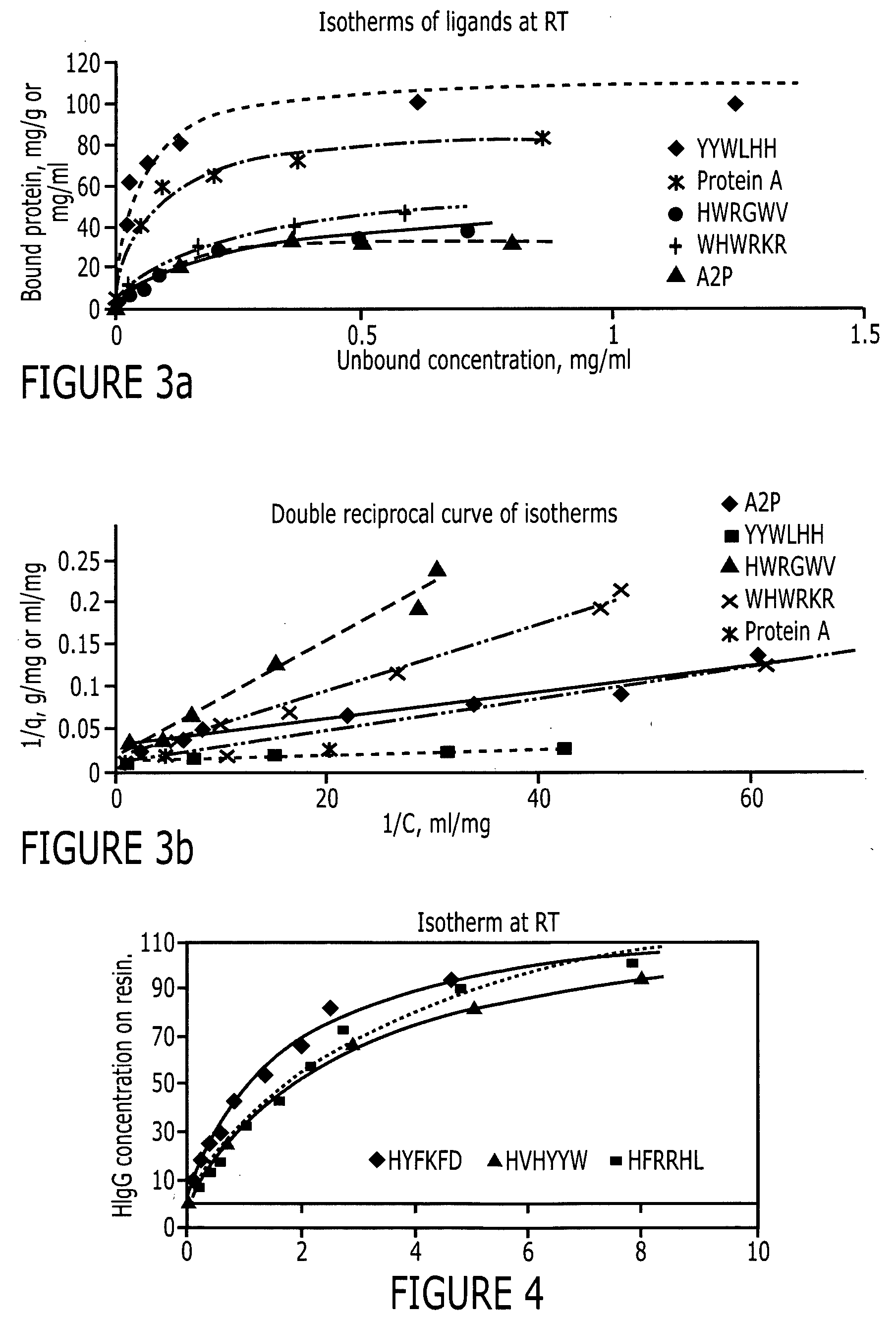
[0084] The peptide libraries used in this study were synthesized by Peptides International (Louisville, Ky.) on Toyopearl AF-amino-650 EC resin (Tosoh Bioscience, Montgomeryville, Pa.). The base resin (FIG. 2) from Tosoh Bioscience is a hydroxylated polymethacrylate amino resin with a pore size of 1000 Å and a particle size of 100-300 μm. The large particle size is particularly advantageous for use in screening since it makes it easier to identify and sequence the particles. Fmoc-L-alanine, a protecting group, was coupled to the amino functionality on the resin. Subsequently, the amino functionalities protected by Fmoc groups were released and used to link to other amino acids. Eighteen of the twenty amino acids (excepting cysteine and methionine) were used in this library synthesis. The amino acid lengths used were six (hexameric library), three (trimeric library) or one (monomeric library) and the peptide density was 400 μmoles / gm. The structure of this library i...
example 2
Positive Controls Used in Screenings
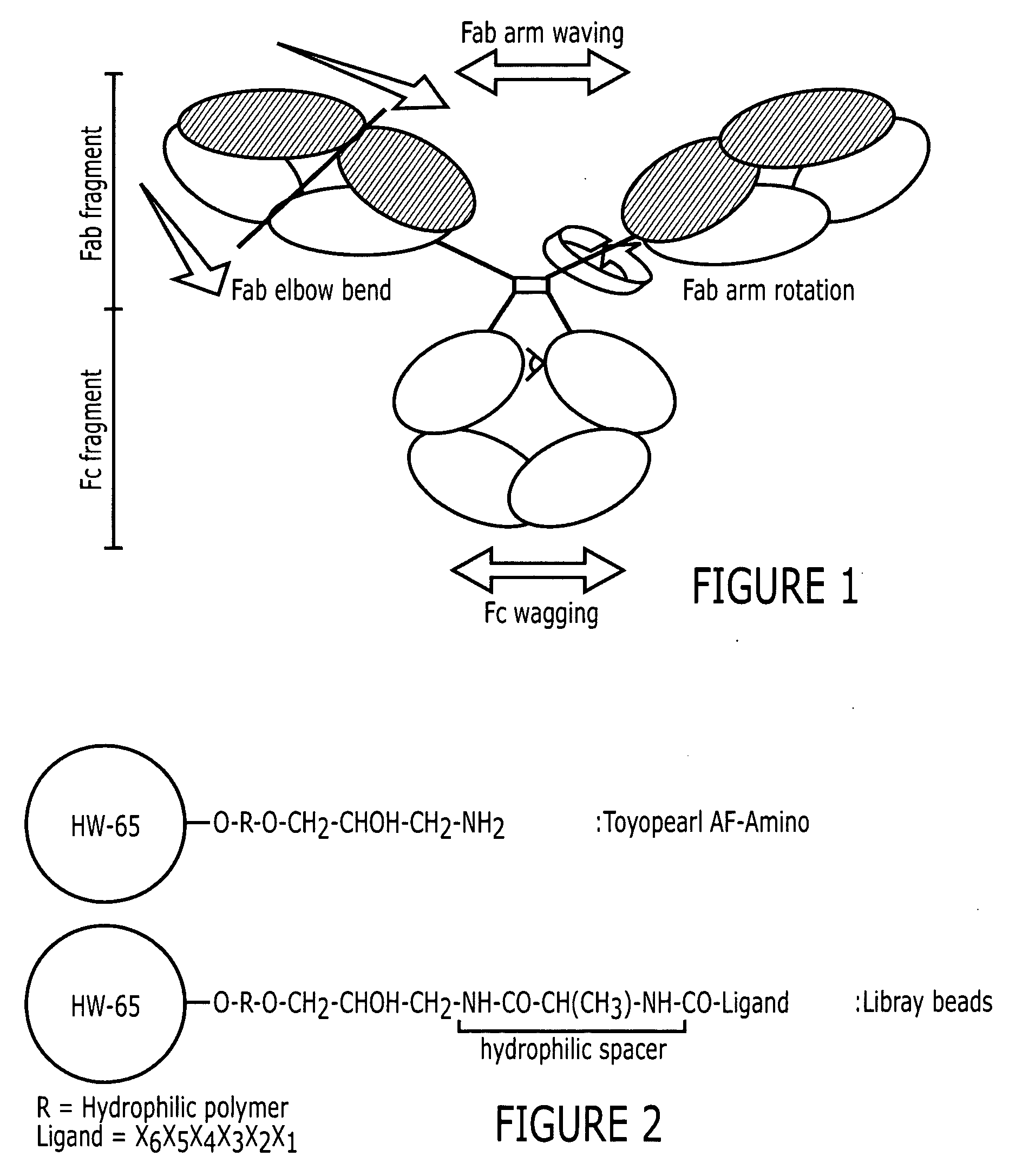
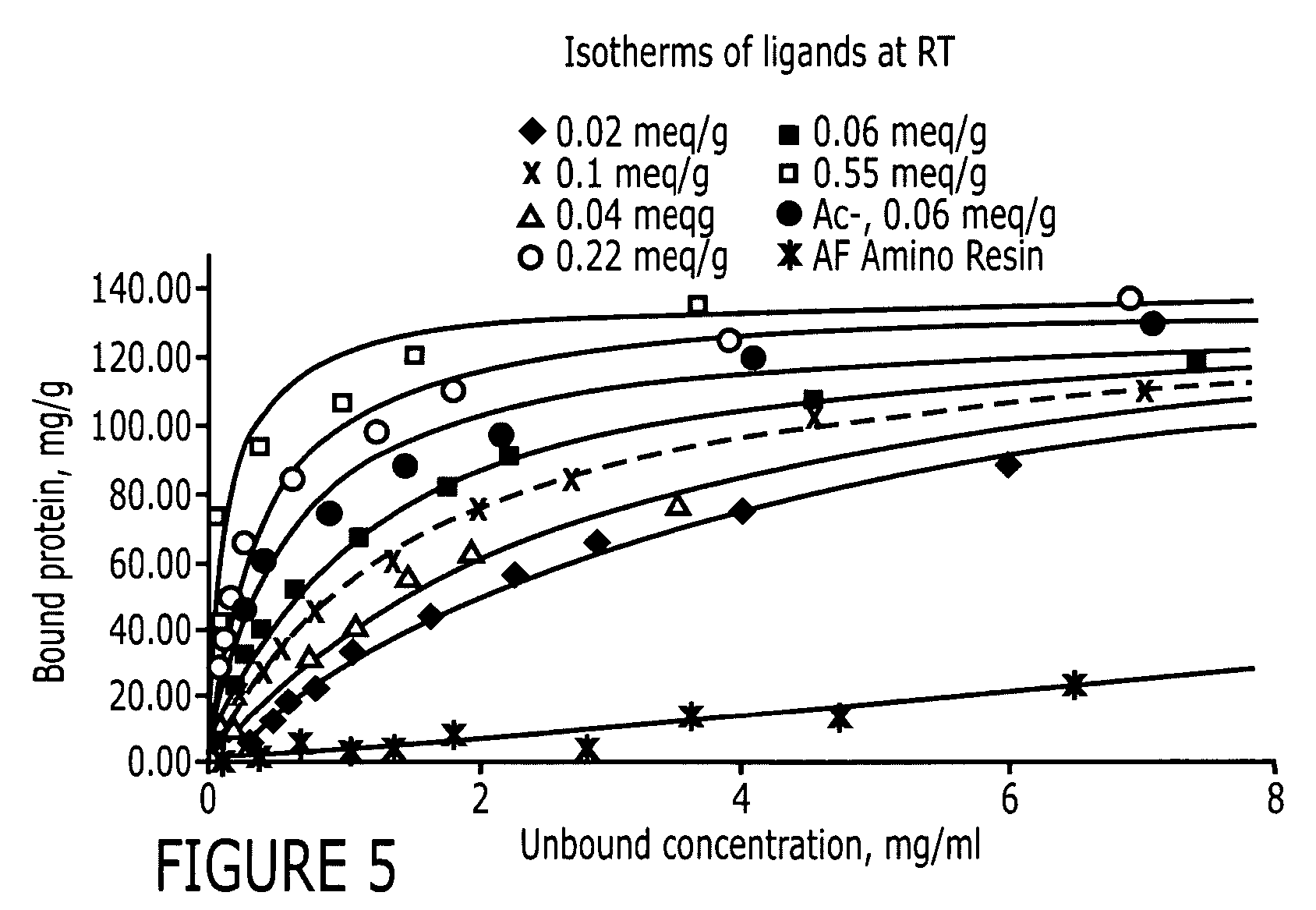
[0086] Two positive controls were used in this study to compare HIgG binding properties. They are protein A Sepharose CL-4B (protein A for short) and MAbsorbent A2P (A2P for short). Protein A Sepharose CL-4B (Amersham Biosciences, Piscataway, N.J.) is protein A immobilized by the CNBr method to 4% cross-linked agarose beads (Sepharose CL-4B) with a mean size of 90 μm. Protein A binds only to the Fc fragment of HIgG 1,2 and 4. MAbsorbent A2P (Prometic Biosciences, Burtonsville, Md.) is made by immobilizing a synthetic triazine based bifunctional ligand onto a 6% cross-linked agarose matrix (PuraBead 6XL) whose mean particle size is around 100 μm. MAbsorbent A2P binds to both Fc and Fab fragment as well as all sub-classes of IgG.
example 3
Radiolabeling of Whole HIgG and Fc Fragment of HIgG
[0087] Radiolabeling process. Fc fragment and HIgG (Bethyl Laboratories, Inc. Montgomery, Tex.) were labeled by reductive methylation (Equation 1, below) utilizing sodium cyanoborohydride (NaCNBH3) and 14C-formaldehyde (H14CHO, PerkinElmer) (Jentoft and Dearborn, 1983). The 14C formaldehyde was added at a 3-fold molar excess over 5% of the total methylation sites (39 total in Fc and more than 85 in intact HIgG). Sodium cyanoborohydride was added at 10× the amount of formaldehyde. The reaction was performed at 4° C. overnight. Following the reaction, the labeled protein was separated from 14C-formaldehyde using an EconoPac 10DG Desalting Column (Biorad, Hercules, Calif.) equilibrated with 0.1 M sodium phosphate, pH 7.0. The radioactivity of each fraction was determined using a Pacard 1500 Tri-Carb Liquid Scintillation Analyzer (Meridan, Conn.) and CytoScint ES scintillation liquid from ICN (Mesa, Calif.). The protein concentration i...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com