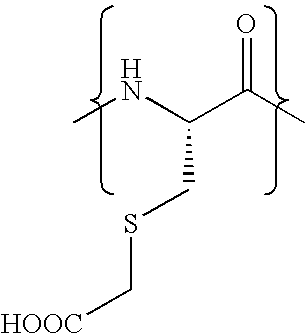
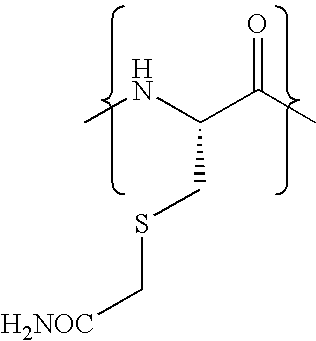
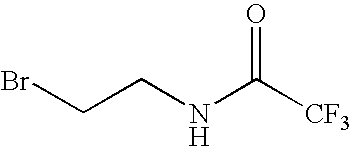
Pseudo-native chemical ligation
a chemical ligation and native technology, applied in the field of pseudo-native chemical ligation, can solve the problems of solid-phase bound products, difficult to obtain high-purity well-defined products, and failure to use recombinant dna techniques
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Synthesis of Synthetic Erythropoiesis Stimulating Protein SEP-0
[0167] A synthetic erythropoiesis stimulating protein (SEP) was synthesized. The sequence of the full-length synthesized protein (designated “SEP-0 (1-166)” is:
(SEQ ID NO:1)APPRLICDSR VLERYLLEAK EAEKITTGCA EHCSLNEKITVPDTKVNFYA WKRMEVGQQA VEVWQGLALL SEAVLRGQALLVKSSQPWψP LQLHVDKAVS GLRSLTTLLR ALGAQKψAISPPDAASAAPL RTITADTFRK LFRVYSNFLR GKLKLYTGEACRTGDR
[0168] where Ψ denotes a non-native amino acid residue consisting of a cysteine that is carboxymethylated at the sulfhydryl group. The SEP-0 protein was synthesized in solution from four polypeptide segments:
Segment SEP-0:1 (GRFN 1711; composed of residues1-32 of SEQ ID NO:1):APPRLICDSR VLERYLLEAK EAEKITTGCA EH-thioesterSegment SEP-0:2 (GRFN 1712, composed of residues33-88 of SEQ ID NO:1):CSLNEKITVP DTKVNFYAWK RMEVGQQAVE VWQGLALLSEAVLRGQALLV KSSQPW-thioester(where Cys33 is Acm protected)Segment SEP-0:3 (GRFN 1713, composed of residues89-116 of SEQ ID NO:1):CPLQLHVDKA VSGL...
example 2
Synthesis of Synthetic Erythropoiesis Stimulating Protein SEP-1-L30
[0179] A second synthetic erythropoiesis stimulating protein (designated SEP-1-L30) was synthesized to contain oxime-forming groups at positions 24 and 126 of SEP-0. These groups were then used to form SEP-1-L30, in which linear (EDA-Succ-)18 carboxylate (EDA=(4,7,10)-trioxatridecane-1,13diamine, also called TTD; Succ=—CO—CH2CH2CO—) polymers have been joined to the protein backbone. The sequence of the full-length SEP-1 (1-166) is:
(SEQ ID NO:2)APPRLICDSR VLERYLLEAK EAEKoxITTGCA EHCSLNEKITVPDTKVNFYA WKRMEVGQQA VEVWQGLLALL SEAVLRGQALLVKSSQPWψP LQLHVDKAVS GLRSLTTLLR ALGAQKψAISPPDAAKoxAAPL RTITADTFRK LFRVYSNFLR GKLKLYTGEACRTGDR
[0180] where Ψ denotes an non-native amino acid residue consisting of a cysteine that is carboxymethylated at the sulfhydryl group, and where Kox denotes a non-native lysine that is chemically modified at the ε-amino group with an oxime linker group coupled to a designated water-soluble polymer ...
example 3
Synthesis of Synthetic Erythropoiesis Stimulating Protein SEP-1-L26
[0189] A third synthetic erythropoiesis stimulating protein (designated SEP-1-L26) was synthesized to contain oxime-forming groups at positions 24 and 126 of SEP-0. These groups were then used to form SEP-1-L26, in which the linear polymers (EDA-Succ)18 carboxylate and (EDA-Succ)6-amide have been joined to the protein backbone through oxime linkages at positions 24 and 126, respectively. The sequence of the full-length SEP-1 (1-166) is:
(SEQ ID NO:2)APPRLICDSR VLERYLLEAK EAEKoxITTGCA EHCSLNEKITVPDTKVNFYA WKRMEVGQQA VEVWQGLALL SEAVLRGQALLVKSSQPWψP LQLHVDKAVS GLRSLTTLLR ALGAQKψAISPPDAAKoxAAPL RTITADTFRK LFRVYSNFLR GKLKLYTGEACRTGDR
where Ψ denotes an non-native amino acid residue consisting of a cysteine that is carboxymethylated at the sulfhydryl group, and where Kox denotes a non-native lysine that is chemically modified at the F-amino group with an oxime linker group coupled to a designated water-soluble polymer th...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| MW | aaaaa | aaaaa |
| MW | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com