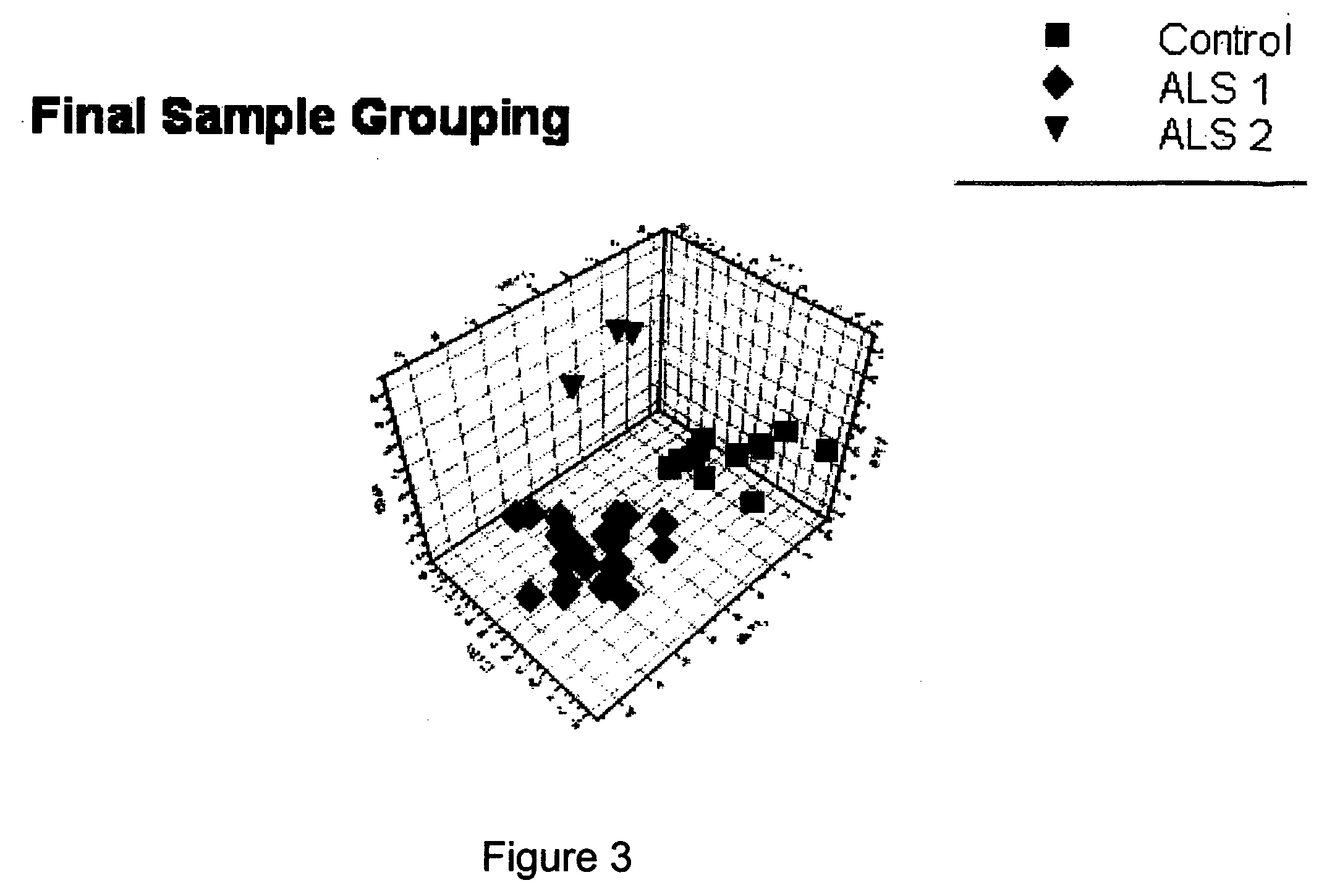
Methods for drug discovery, disease treatment, and diagnosis using metabolomics
a technology of metabolomics and drug discovery, applied in the direction of biochemistry apparatus and processes, instruments, ict adaptation, etc., can solve the problems of expensive and time-consuming procedures, and achieve the effect of accelerating the process of identifying genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Method for Obtaining a Small Molecule Profile of a Cellular Compartment
[0271] Method for Obtaining a Small Molecule Profile of Mitochondria using HPLC
[0272] The following method demonstrates how small molecules are isolated from mitochondria for methods as described herein.
[0273] Mitochondrial Isolation
[0274] Mitochondria from a mammalian source are isolated by differential centrifugation in 140 mM KCI and 20 mM Hepes, pH 7.4. Following the final wash, mitochondria are resuspended in the same buffer and aliquots are quick frozen in liquid nitrogen. Protein determinations are carried out by Lowry using the Sigma Protein Assay Kit P5656).
[0275] Other mitochondrial samples are purified using a modified version of a published protocol (Rigobello et al. (1995) Arch. Biochem. Biophys. 319, 225-230). Mammalian liver mitochondria are obtained after decapitation of the subjects. The livers are dissected out and are placed in an ice-cold solution containing 250 mM mannitol, 75 mM sucrose...
example 2
Method for Analyzing Metabolic Disorders Using Small Molecule Profiles
[0284] Method for Analyzing Differences in Small Molecule Profiles of Genetically Altered Mice After Short Term Diet Variations
[0285] 15 female C57B1 / 6J ob / ob mice and lean littermate controls (15 female C57B1 / 6J ? / +) and 15 male C57B1 / Ks db / db mice and lean littermate controls (15 male C57B1 / ks + / +) are obtained from Jackson labs at 4.5 weeks of age, and are housed individually on normal mouse chow (West, D. B., 1992, Am. J. Physiol 262:R1025-R1032) for 1 week prior to the initiation of the study. The four groups of 15 mice each are then sacrificed by CO2 euthanasia and tissues are then collected. Body weight (grams) of the four groups of mice at the time of sacrifice are measured. Small molecule profiles from cells from the hypothallumus are then obtained from each group of mice and compared.
[0286] The mice (normal, lean, ob / ob, db / db, and / or tub / tub) are fed normally prior to the initiation of the experimen...
example 3
Comparison of Small Molecule Profiles of Different Types of Immune Cells
[0289] Method for Comparing the Small Molecule Profiles of TH1 and TH2 Cells
[0290] The transgenic T cell example is used to identify cellular small molecules present in TH2 cells. The identified small molecules are be present in different amounts in TH2 cells compared to TH1 cells.
[0291] Transgenic Mice
[0292] Naive CD4+ cells are obtained from the spleens and / or lymph nodes of unprimed transgenic mouse strains harboring a T cell receptor (TCR) recognizing ovalbumin (Murphy et al., 1990, Science 250:1720).
[0293] Ova-Specific Transgenic T Cells
[0294] Suspensions of ova-specific T cells are co-cultured with stimulatory peptide antigen and antigen presenting cells essentially as described in Murphy et al. (Murphy et al., 1990, Science 250:1720). Briefly, 2-4×106 T cells are incubated with approximately twice as many TA3 antigen presenting cells in the presence of 0.3 μM Ova peptide. TH1 cultures may contain ap...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelengths | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com