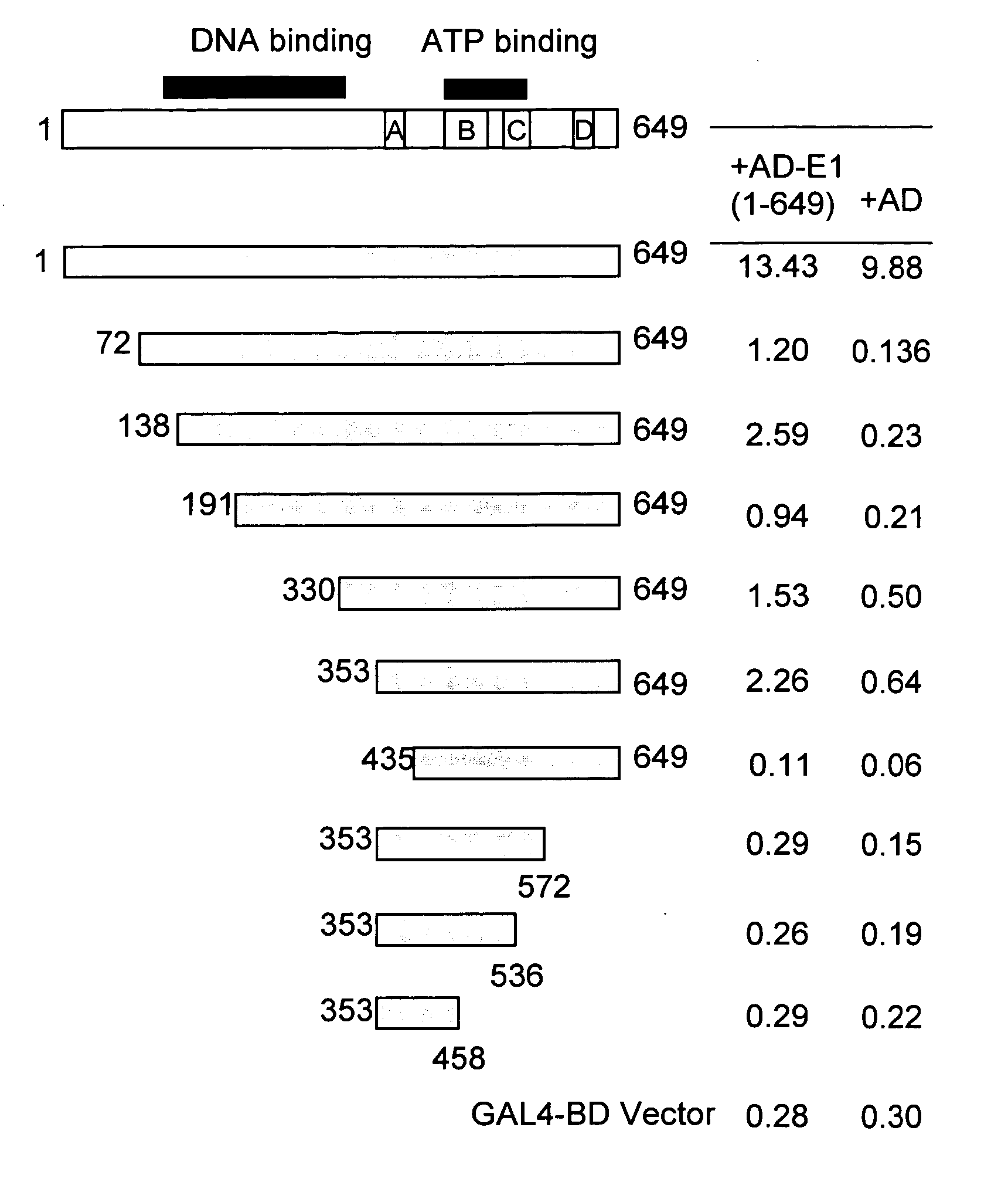
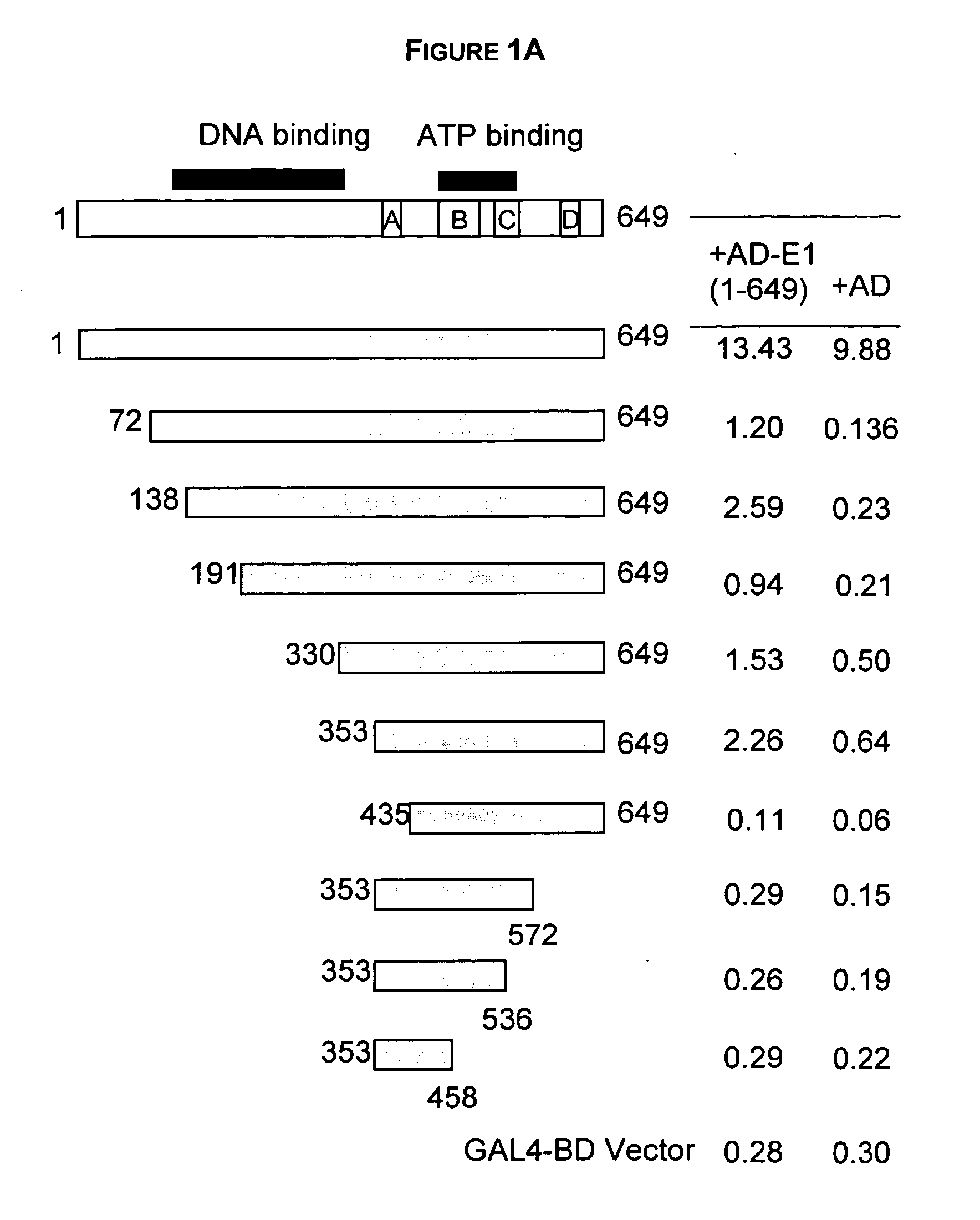
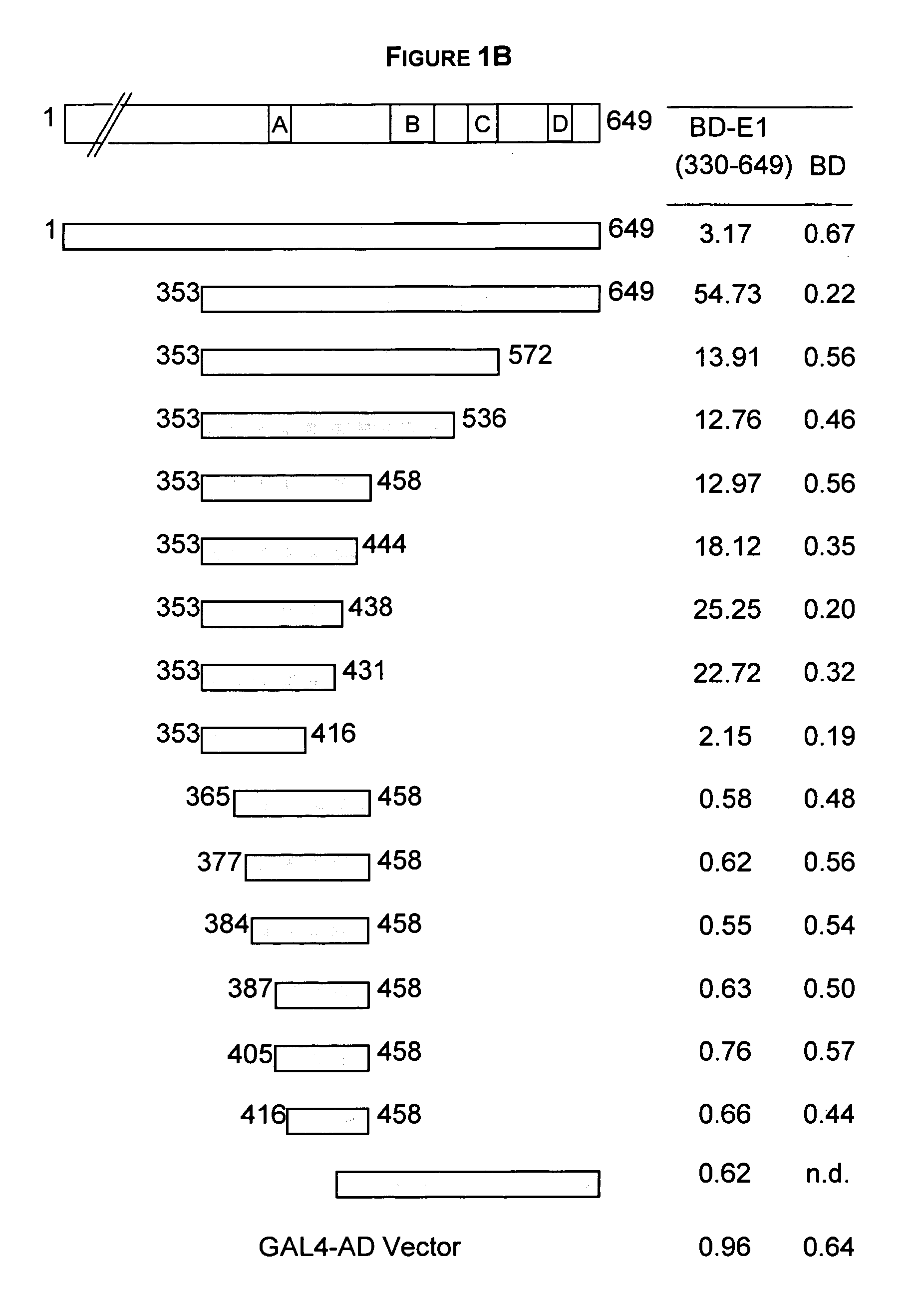
Regions of papilloma virus E1 helicase involved in E1 oligomerization
a helicase and papilloma virus technology, applied in the field of papilloma virus e1 helicase involved in e1 oligomerization, can solve the problems of no effective antiviral treatment for hpv infection, no treatment of all viral particles, and high cost incurred or uncomfortable side effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Yeast Strain, Media, and Genetic Methods
[0133]Saccharomyces cerevisiae strain Y153 (MATa leu2-3, 112 ura3-52 trp1-901 his3-Δ200 ade2-101 gal4Δgal80Δ URA3::GAL-lacZ LYS::GAL-HIS3) was used for yeast two-hybrid analysis (Durfee et al., 1993, Genes. Dev. 7:555-569). Transformation of yeast strain Y153 was performed using the LiAc method essentially as described in the Clontech Matchmaker Library Protocol. Cells that were co-transformed with a combination of two plasmids were selected at 30° C. for 3 to 5 days on SD medium (described by Sherman et al. 1979, Methods in Yeast Genetics, Cold Spring Harbor, N.Y.) lacking leucine and tryptophan but supplemented with the other required amino acids.
example 2
β-Galactosidase Assays
[0134] Transformed yeast cells were pre-grown in liquid SD medium lacking leucine and tryptophan and then used to inoculate YPD (Sherman et al. Supra) cultures. These cultures were grown at 30° C. until they reached an optical density of approximately 0.6 at 600 nm (OD600). Cells were then harvested, washed and permeabilized by two cycles of freezing (liquid nitrogen) and thawing. β-galactosidase activity was then measured spectrophotometrically (at 578 nm) using the substrate chlorophenyl-red-β-D-galactopyranoside (CRPG, Boehringer Mannheim) as described in the Clontech Matchmaker Library Protocol. Enzymatic activity was calculated using the equation: Miller unit=(1000×OD578) / (elapsed min×1.5 ml culture×OD600).
example 3
Plasmid Constructions
A. Plasmids for In-Vitro Transcription / Translation
[0135] The constructs and the primers for amplification are summarized in Table 1.
[0136] Plasmids used for synthesis of HPV-11 E1 and E2 in vitro were derived either from pCR3 (Invitrogen, CA) or from pTM1 (obtained from Bernard Moss, NIH). In these plasmids, the encoded protein can be expressed in vitro from the T7 promoter located upstream of the open reading frame (ORF). When used in a coupled transcription / translation system (TNT Coupled Reticulocyte Lysate System, Promega), plasmids derived from pTM1 directed the synthesis of higher levels of proteins. Presumably, it is because this plasmid encodes the EMCV IRES (encephalomyocarditis virus internal ribosome entry site), which stimulates translation (data not shown).
[0137] To construct pCR3-E1 and pCR3-E2, the entire HPV-11 E1 and E2 ORFs were amplified separately by polymerase chain reaction (PCR), though any method capable of amplifying DNA is suitable...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperatures | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com