Oligonucleotides and assemblies thereof useful in the detection of the presence or absence of target nucleic acid sequences in a sample
a technology of oligonucleotide and target nucleic acid sequence, applied in the field of oligonucleotides, can solve the problems of poor binding stability and selectivity, limited use of linear oligonucleotide probes, and inability to enable signal amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Materials and General Methods
[0132] Oligonucleotides:
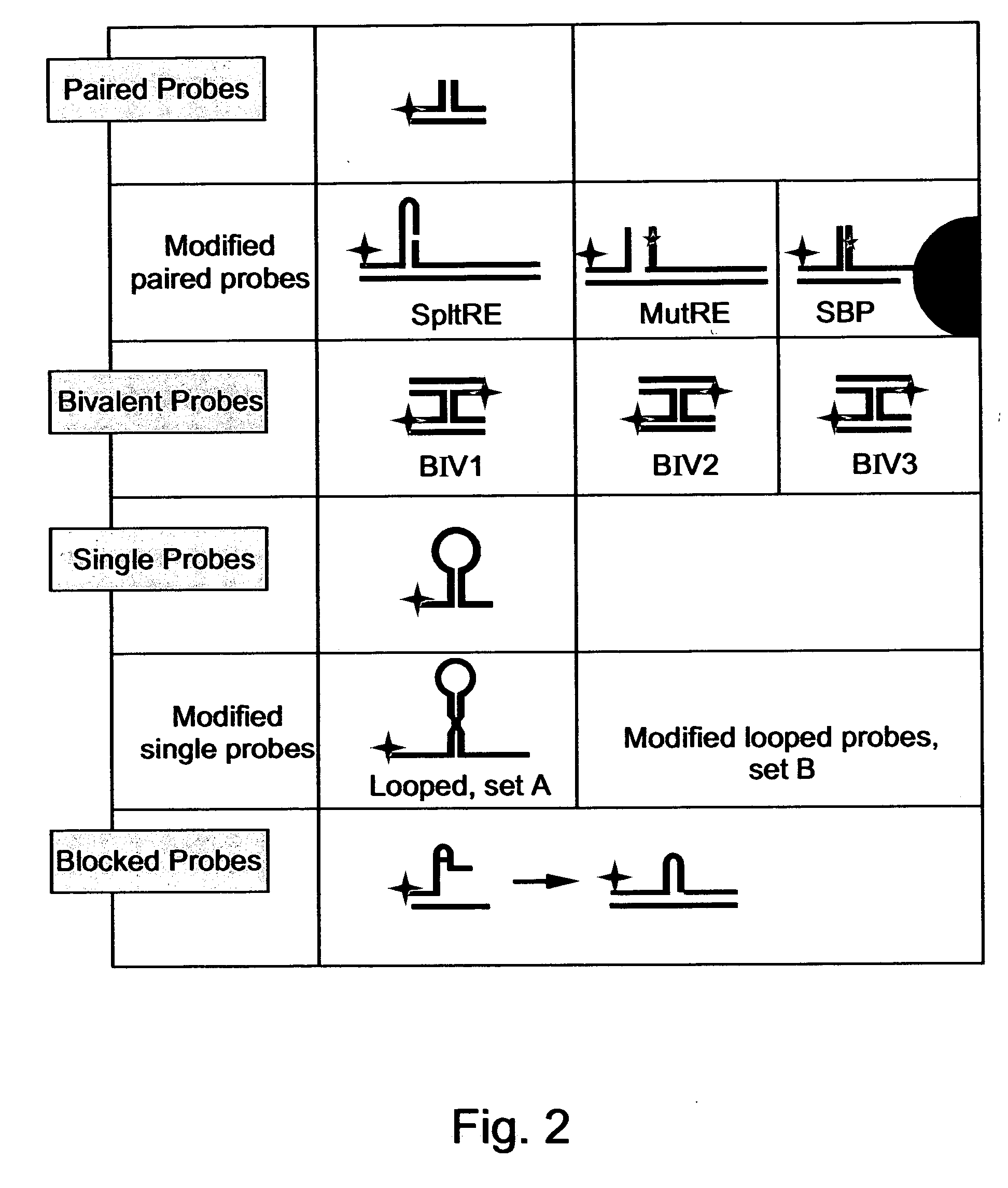
[0133] Following careful design, the oligonucleotides and oligonucleotide assemblies described hereinbelow were acquired from Biotechnology General Ltd., Israel or Genset, France. The prediction of structure and thermodynamic stability of the oligonucleotides, at different assay conditions, was performed using the Gene runner software, version 3.00. FIG. 2 summarizes the secondary structures and features of all of the oligonucleotides synthesized and tested while reducing the present invention to practice.
[0134] PCR Reactions:
[0135] PCR reactions were conducted using the Programmable Thermal Controller PTC-100.TM. (MJ Research, Inc.). The DNA utilized as template in the PCR reactions was prepared from whole-cell lysate of human embryo fibroblast (HEF) cells infected with cyto-megalo virus (CMV, ATCC strain AD169).
[0136] Restriction Endonucleases and Carrier DNA:
[0137] Restriction enzymes used for template DNA and oligonucleotide p...
example 2
Target Nucleic Acids
[0149] CMV-DNA Preparations:
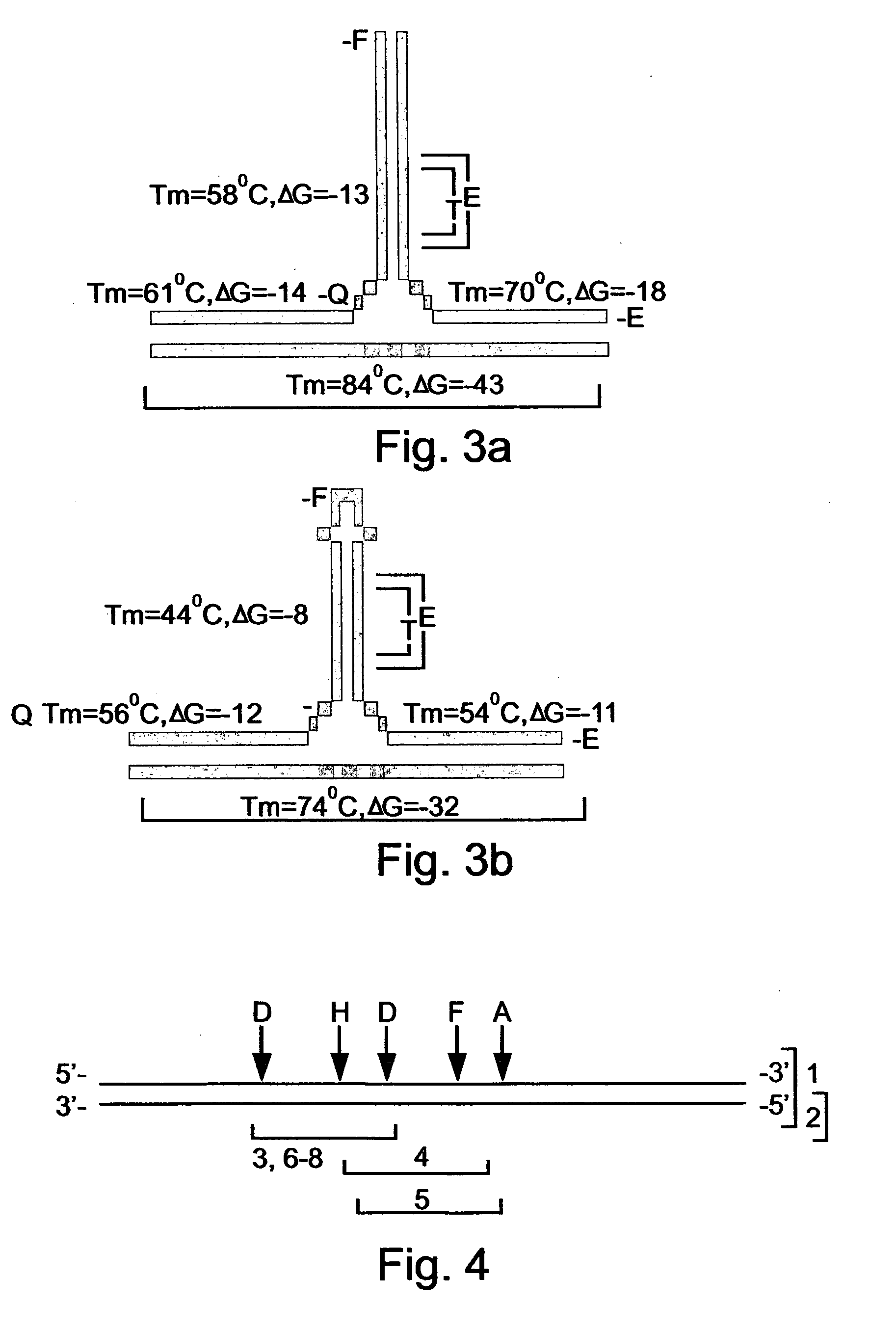
[0150] A 263 base pair fragment (SEQ ID NO:1) derived from the CMV genome (VRL Accession No. X17403) was used as a template for Various CMV-DNA preparations which were used as a target DNA sequence (FIG. 4). The main features of the various preparations are listed in Table 1 and are further detailed hereinbelow.
[0151] PCR amplified double stranded CMV-DNA (p.CMV-263.ds): This unique CMV fragment was amplified using 5'-AGACCTTCATGCAGATCTCC-3' (sense CMV-PCR primer, SEQ ID NO:2) as a sense primer and 5'-GGTGCTCACGCACATTGATC-3' (antisense CMV-PCR primer, SEQ ID NO:3) as an antisense primer, along with a DNA preparation from whole-cell lysate of CMV-infected HEF cells as a PCR template. The PCR reaction included a first denaturing step of 5 minutes at 94.degree. C., followed by thirty cycles of 1 minute at 94.degree. C.; 30 seconds at 58.degree. C.; 30 seconds at 72.degree. C. and a final extension step of 5 minutes at 72.degree. C.
1TABLE ...
example 3
Bi-molecular-Paired Probes
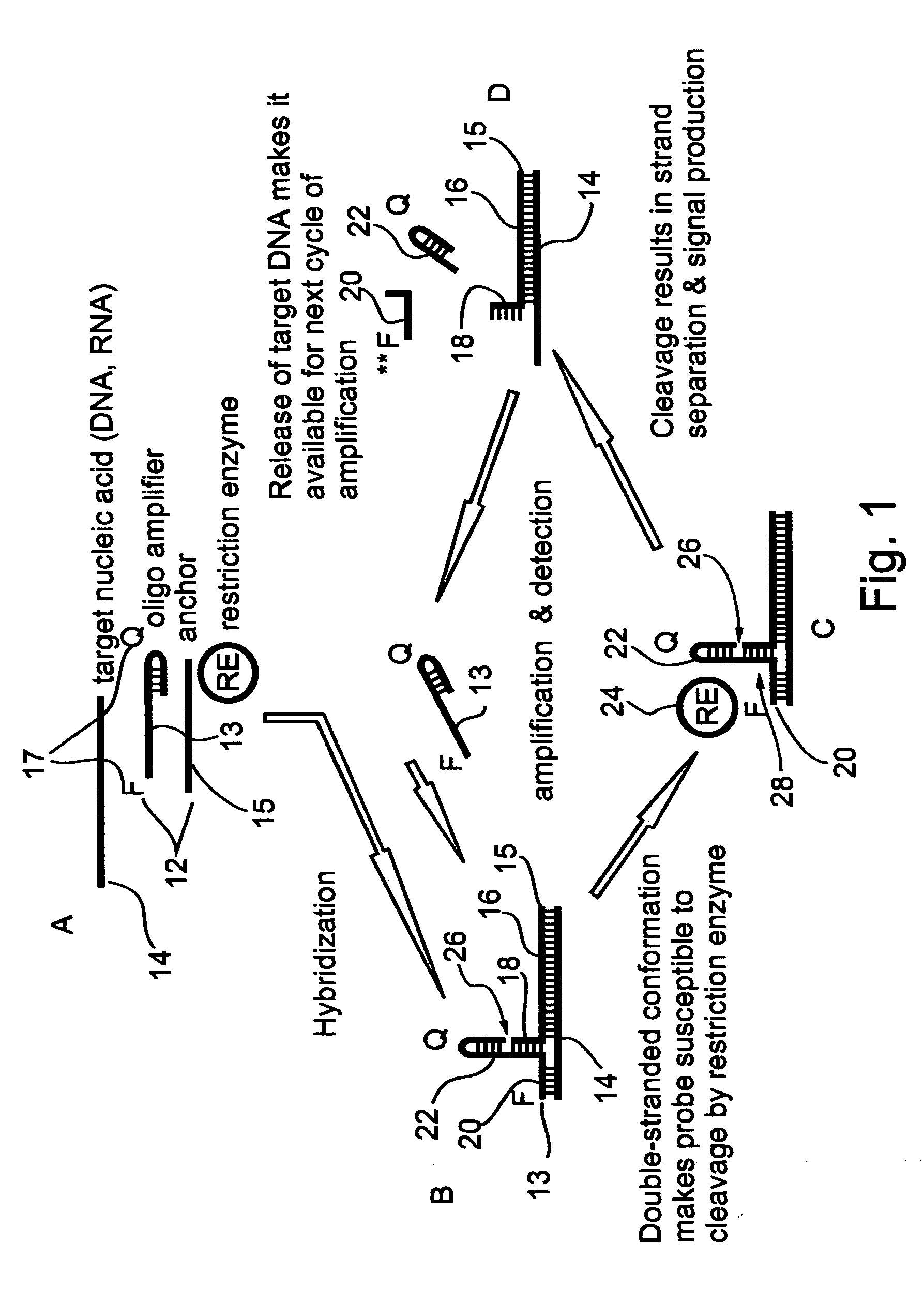
[0157] Paired probes--First generation: In paired oligonucleotide probes (bi-molecular oligonucleotide assemblies) each oligonucleotide member of the pair contains an arm which is designed to specifically recognize a portion of the target sequence. However, each arm is preferably selected sufficiently short so as to prohibit the formation of a stable hybrid with the target sequence on its own. The concomitant hybridization of the arms of both oligonucleotide members of the pair to the target DNA allows the formation of a double stranded stem (11-18 bp long) between the two oligonucleotide members. This stem is essential for the stabilization of the hybridization between the arms of both oligonucleotide members and the target DNA. In addition, this stem is designed to provide cleavable restriction sites which are formed as a result of the stem structure formation. The specifics are further detailed hereinbelow.
[0158] Effects of stem and arms regions on hybri...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com