Membrane chip for detecting 45 gene locus of mitochondria diabetes
A gene locus and diabetes technology, which is applied in the field of membrane chip detection of 45 gene loci of mitochondrial diabetes, can solve the problems of inability to systematically detect mitochondrial mutation sites, no control on the chip, and long time-consuming experiments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Sample Handling and Labeling
[0055] (1) Fresh whole blood samples
[0056] The modified NaI method (Yu Hong, Peng Fangfang. Experimental Techniques of Medical Biochemistry and Molecular Biology, 1st edition, Wuhan University Press, 2003) was used to extract DNA templates.
[0057] (2) blood stain specimen
[0058] Take 1.5×1.5cm 2 For large or small blood stains, immerse in 1ml double distilled water, shake and mix well, centrifuge at 12,000rpm for 3min, remove the supernatant, add double distilled water to wash once, then add 200μl 5% Chelex-100, incubate at 56°C for 20-30min, then boil After 8 minutes, centrifuge at 10,000 rpm for 3 minutes, and take the supernatant for PCR amplification.
[0059] (3) hairball specimen
[0060] Cut off 3 to 4 hairs at a distance of 3 to 4 mm from the root, soak the hair ball in Chelex-100 solution, and keep it warm at 56°C for 6 to 10 hours. Oscillate for 5-10s. After boiling at 100°C for 10 minutes, centrifuge at 10,000 r / min...
Embodiment 2
[0063] chip fabrication method
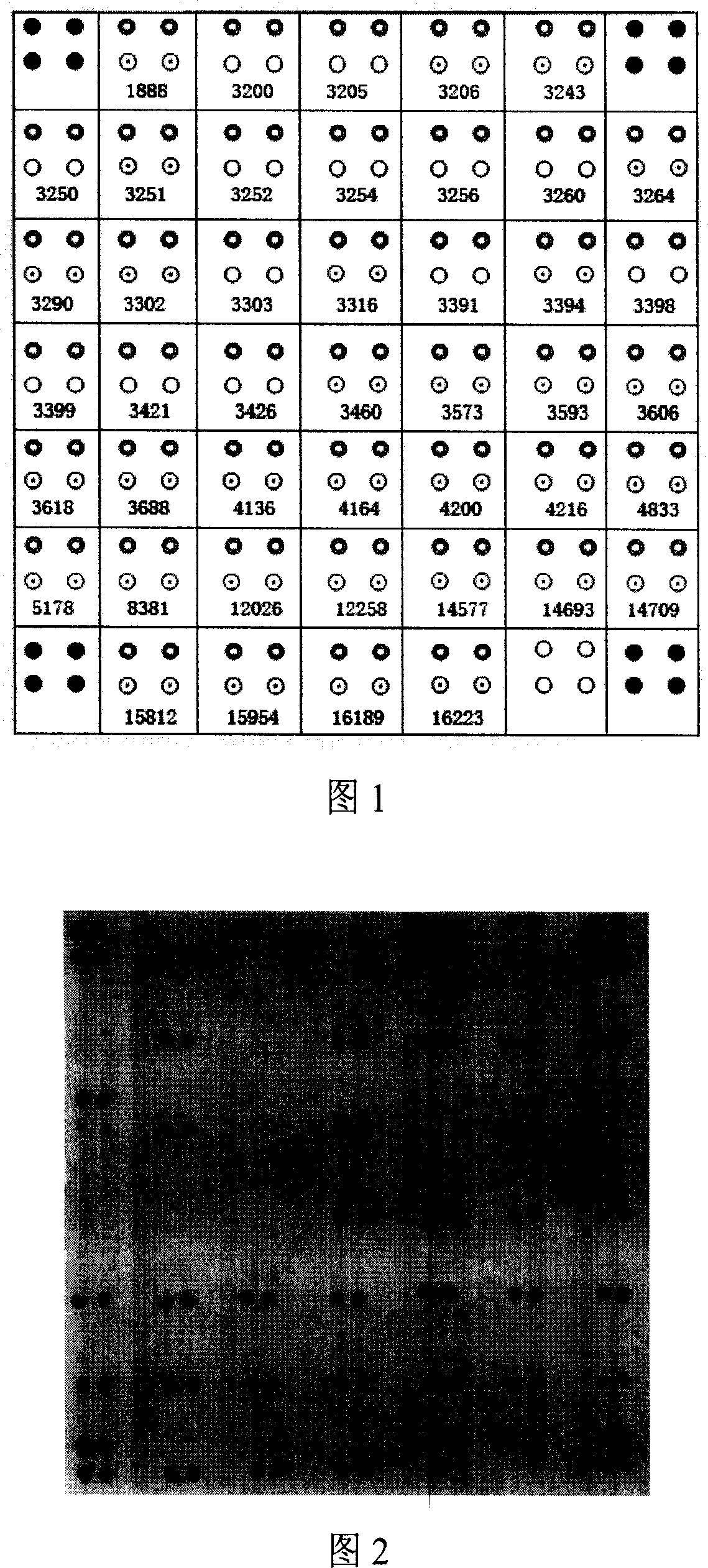
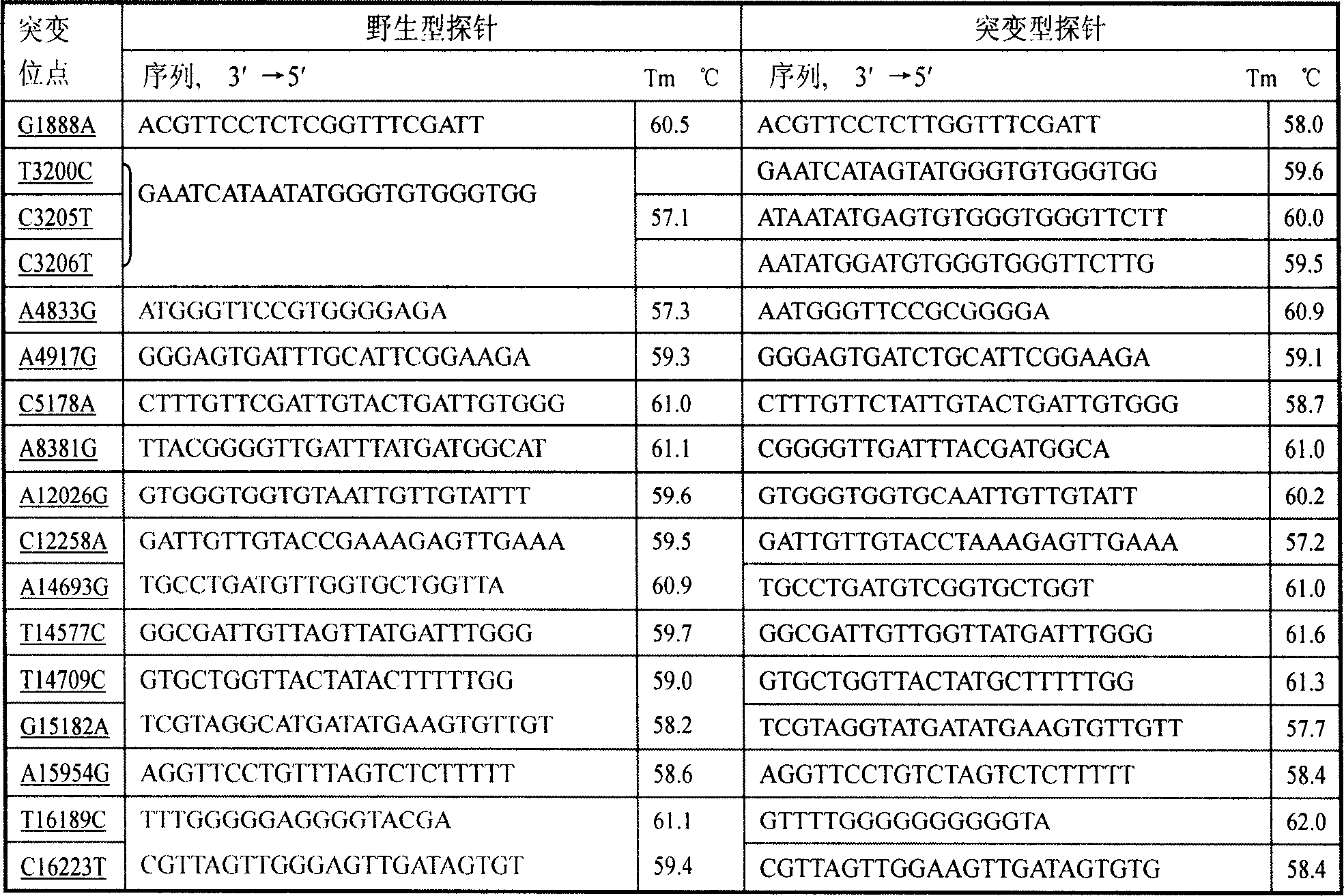
[0064] Nylon membrane into DdH 2 Soak in O for 10 minutes, immerse in 5×SSC-Buffer for 30 minutes (20×SSC-Buffer: 8.766g sodium chloride, 4.412g sodium citrate, distilled water to 50ml, adjust pH to 7.5 with sodium hydroxide), and air dry naturally; 90 amino-modified wild-type and mutant probes at each site were diluted with 5×SSC-Buffer to a probe solution with a final concentration of 25 μm, arranged in a 384-well plate according to the spotting matrix (Figure 1), and used 478A Chip spotting instrument (U.S. UVP Company) dot printing; 8min cross-linking and fixing by ultraviolet light irradiation; store at 4°C for future use.
Embodiment 3
[0066] Hybridization and immunochromogenic detection of mutations
[0067] Preheat the hybridization solution (Roche) at 42°C, immerse the membrane chip in the pre-hybridization solution, and pre-hybridize for 30 minutes to block the non-specific binding of the substrate; mix the biotin-labeled ssDNA with the preheated hybridization solution at a ratio of 1:10, and hybridize at 42°C 2h. Take out the membrane chip, wash and develop color according to the following steps.
[0068] (1) Wash at room temperature (25°C-30°C) with 2×SSC-0.1% SDS (sodium dodecylsulfonate, Sigma) for 5 min×2 times, and 0.5×SSC-0.1% SDS for 15 min.
[0069] (2) Maleic acid Buffer-0.3% Tween20 elution 5min (maleic acid Buffer: 2.32 grams of maleic acid (maleic acid), 1.7532 grams of sodium chloride, dissolved in 200ml of double distilled water, adjusted with sodium hydroxide pH is 7.5; Tween20, Sigma company).
[0070] (3) Block with 1×blocking solution for 30 min (1×blocking solution: 9ml maleic acid...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com