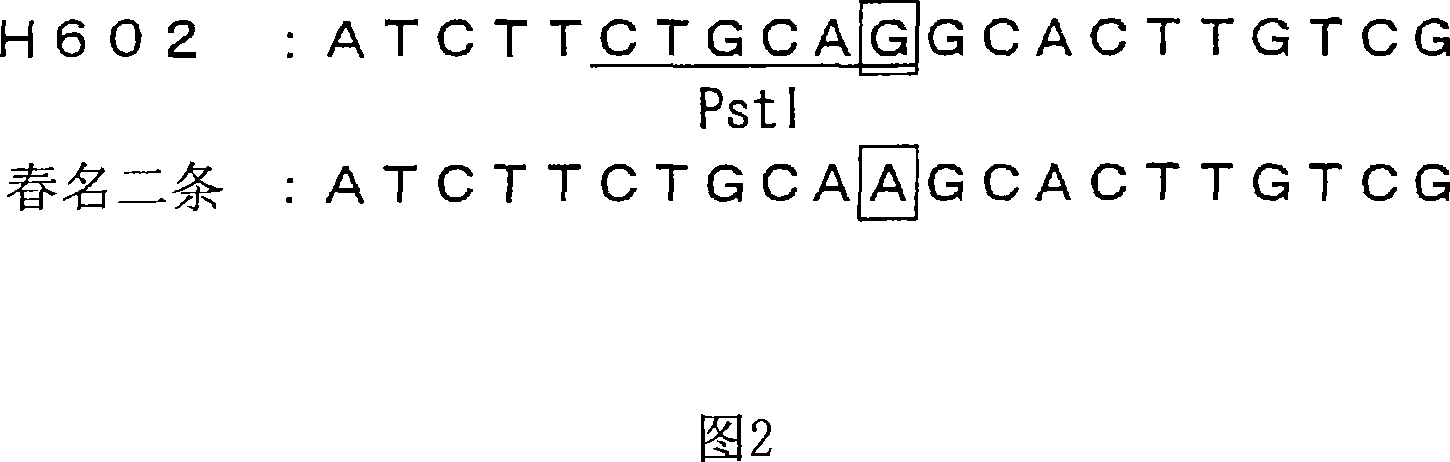
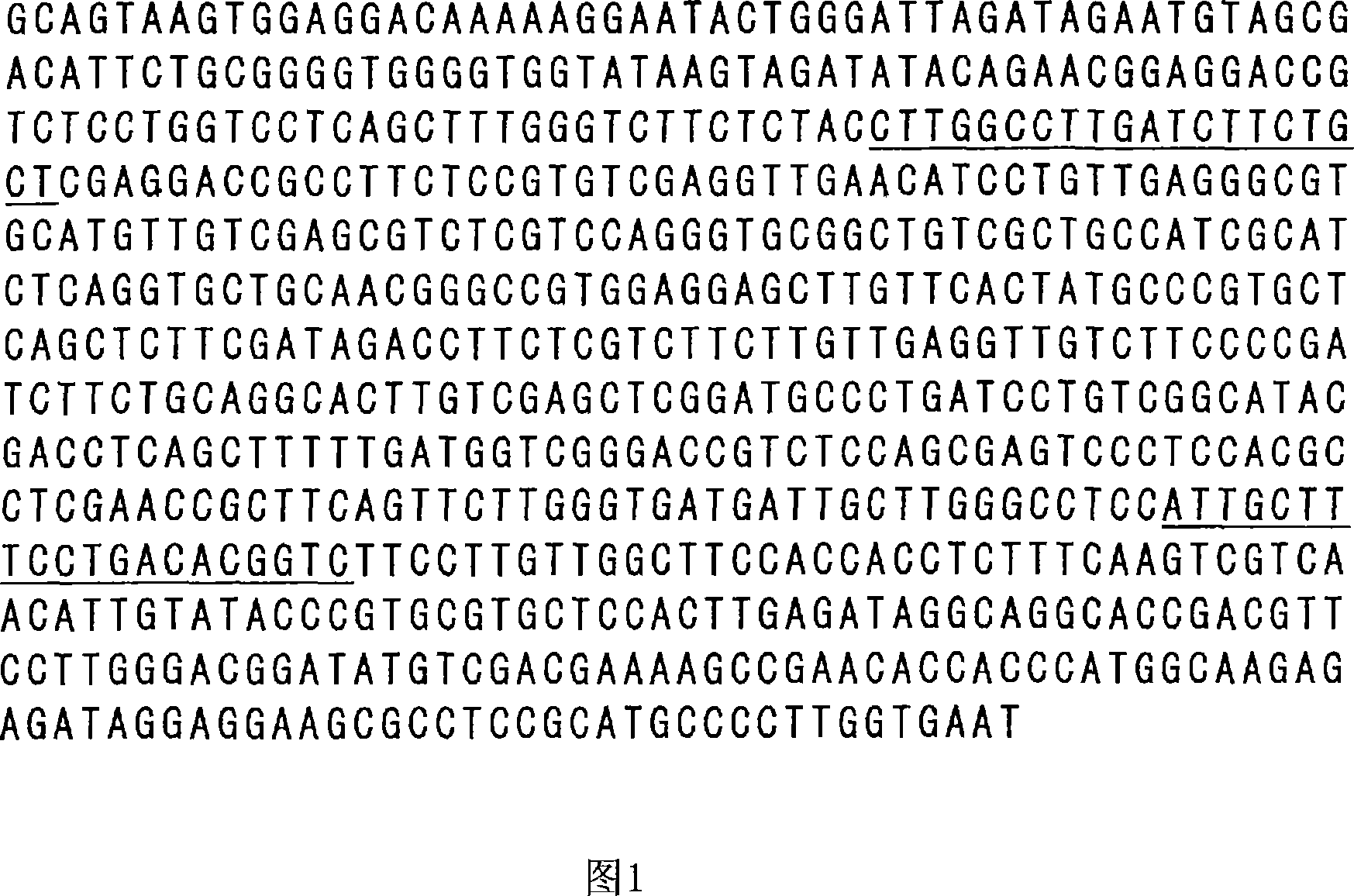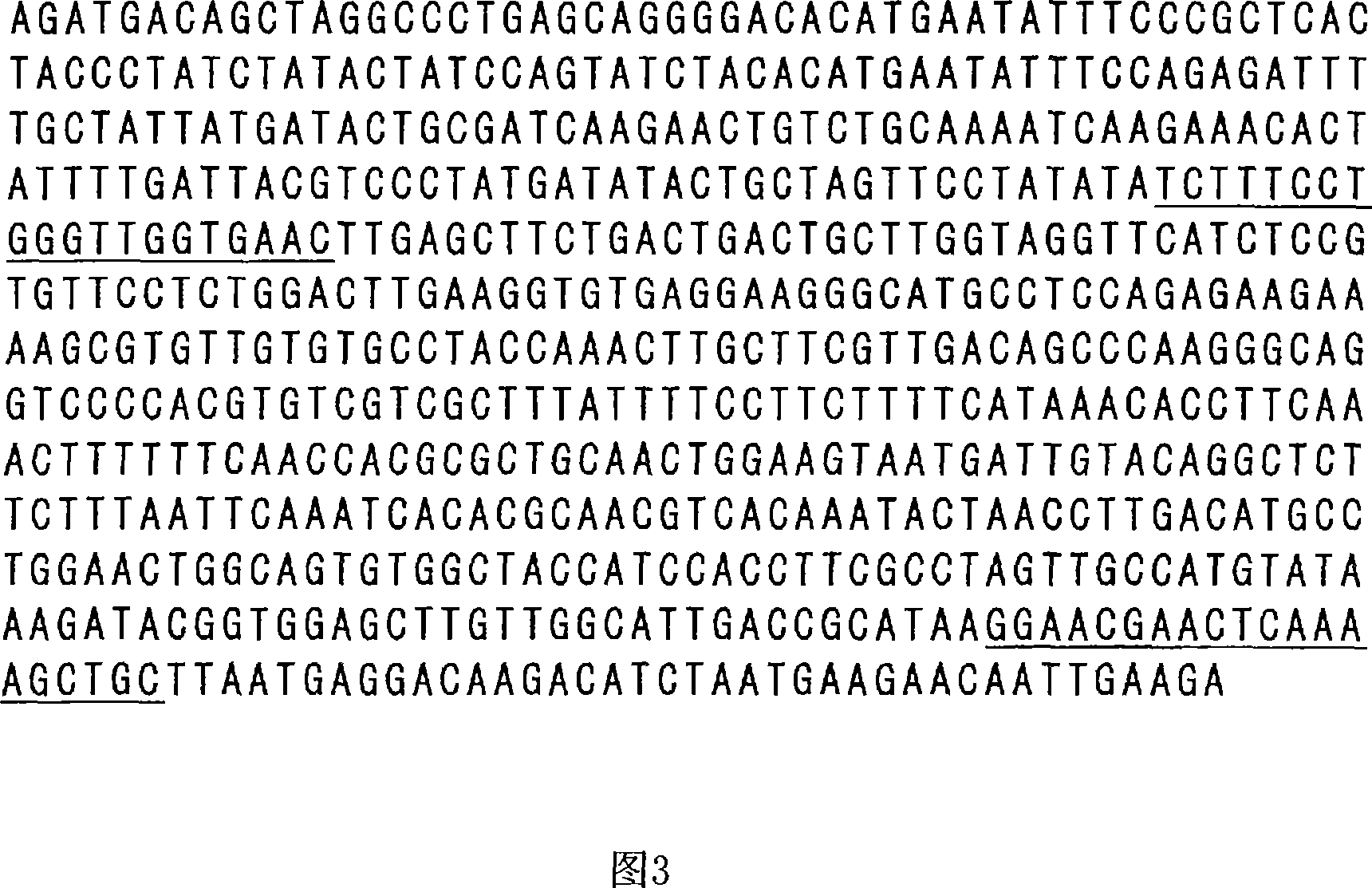Genetic marker linked to gene locus involved in barley resistance to yellow mosaic disease and use thereof
A technology of genetic markers and gene loci, applied in the fields of application, angiosperms/flowering plants, botanical equipment and methods, etc., can solve problems such as difficult to identify and select genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0221] [Example 1: QTL Analysis on Barley Yellow Mosaic Resistance]
[0222] In the algorithm of QTL analysis, simple interval mapping (simple interval mapping, hereinafter referred to as "SIM") and composite interval mapping (hereinafter referred to as "CIM") are used, and the analysis software uses MAPMAKER / QTL and QTL Cartographer performed. The threshold value of the LOD score was set to 2, and when the LOD score exceeded 2, it was estimated that the QTL existed at the position with the largest LOD within the two marker intervals.
[0223] [result]
[0224] Table 1 shows the results of the DHHS group, Table 2 shows the results of the RI1 group, and Table 3 shows the results of the RI2 group. It should be noted that the "group" in the above Tables 1 to 3 refers to the "name of the group for which QTL analysis has been performed", "character" refers to the "character", "chromosome" refers to the "chromosome where the genetic marker is located", and " Marker interval (M1-A...
Embodiment 2
[0255] [Example 2: Detection of genetic marker "FEggaMtgg116"]
[0256] (method)
[0257] 50 ng of genomic DNA of barley to be tested was double-digested with 1.5 U each of EcoRI (manufactured by TAKARABIO) and MseI (manufactured by NEW ENGLAND Biolabs) in a total of 25 μl of reaction system at 37° C. for 12 hours. On the DNA after the restriction endonuclease treatment, with 5 μ M EcoRI linker (base sequence shown in SEQ ID NO: 49 and SEQ ID NO: 50) and 50 μ M MseI linker (base sequence shown in SEQ ID NO: 47 and SEQ ID NO: No. 48) Ligation was performed at 37° C. for 3 hours with 25 U of T4 ligase (manufactured by TAKARABIO). Pre-amplification (preliminary amplification) was performed on the ligated DNA fragments using EcoRI universal primer (base sequence shown in SEQ ID NO: 52) and MseI universal primer (base sequence shown in SEQ ID NO: 51). In a 0.07 mg / ml preamplification reaction solution, an amplification reaction was performed using an EcoRI selection primer having...
Embodiment 3
[0264] [Example 3: Detection of genetic marker "FEgggMcaa585"]
[0265] The method is to perform an amplification reaction (this amplification) using an EcoRI selection primer having the nucleotide sequence shown in SEQ ID NO: 38 and a MseI selection primer having the nucleotide sequence shown in SEQ ID NO: 37. , using the same method as that shown in the above-mentioned embodiment 2.
[0266] (result)
[0267]Figure 13 shows the results of electrophoresis of the amplified fragments obtained by the above amplification reaction. The left and right lanes in Figure 13 represent size markers, and the second lane from the left lane in the same figure shows the results of the above-mentioned AFLP detection on the barley variety Russia6 resistant to barley yellow mosaic disease, and the second lane from the left end lane in the same figure Lane 3 shows the results of the above-mentioned AFLP detection on the barley variety H.E.S.4 suffering from barley yellow mosaic disease. The o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com