Humanized CTLA-4 single chain antibody and human perforin path formed peptide P34 recombinant immunotoxin
A CTLA-4 and single-chain antibody technology, applied in the field of fusion proteins, can solve the problems of strong immunogenicity, low internalization steps, and low efficiency of action, and achieve the effects of weak antigenicity, high efficiency, and low efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] 1. Cloning of guide fragment gene
[0054] The amplified template is the pCANTAB 5E plasmid (GE Company) loaded with the Anti-CTLA-4-hS83 gene, and the amplified template can also be prepared according to the method described by Pistillo et al. (Pistillo MP et al, Molecular characterization and applications of recombinant scFv Abs to CTLA-4 co-stimulatory molecule. Tissue Antigens, 2000, 55: 229-38).
[0055] Primers were designed according to the gene sequence of Anti-CTLA-4-hS83:
[0056] P1 5'-CAGTGAGCTCATGGCCGAGGTGCAGCTGG-3'
[0057] P2 5'-AGTCAAGCTTACCTAGGACGGTCAGCTTGG-3'
[0058] High-fidelity Platimum Pfx DNA polymerase (Invitrogen, USA) was used for PCR amplification. The PCR amplification conditions were as follows: 94°C for 10 min, 94°C for 45 sec, 50°C for 1 min, 68°C for 2 min, a total of 30 cycles. A guide fragment gene of about 750bp was amplified by PCR, and the corresponding restriction site was introduced.
[0059] The PCR product was purified by a ...
Embodiment 2
[0078] 1. Cloning of hS83 gene
[0079] Using the Anti-CTLA-4-hS83 plasmid as a template, design primers based on its gene sequence:
[0080] P5 5'-CAGTGAGCTCATGGCCGAGGTGCAGCTGG-3'
[0081] P6 5'-CAGTGTCGACTCAACCTAGGACGGTCAGCTTGG-3'
[0082] PCR was performed with high-fidelity Platimum Pfx DNA polymerase, and the PCR conditions were as follows: 10 min at 94°C, 45 sec at 94°C, 1 min at 50°C, 2 min at 68°C, a total of 30 cycles. A guide fragment gene of about 750bp was amplified by PCR, and the corresponding restriction site was introduced.
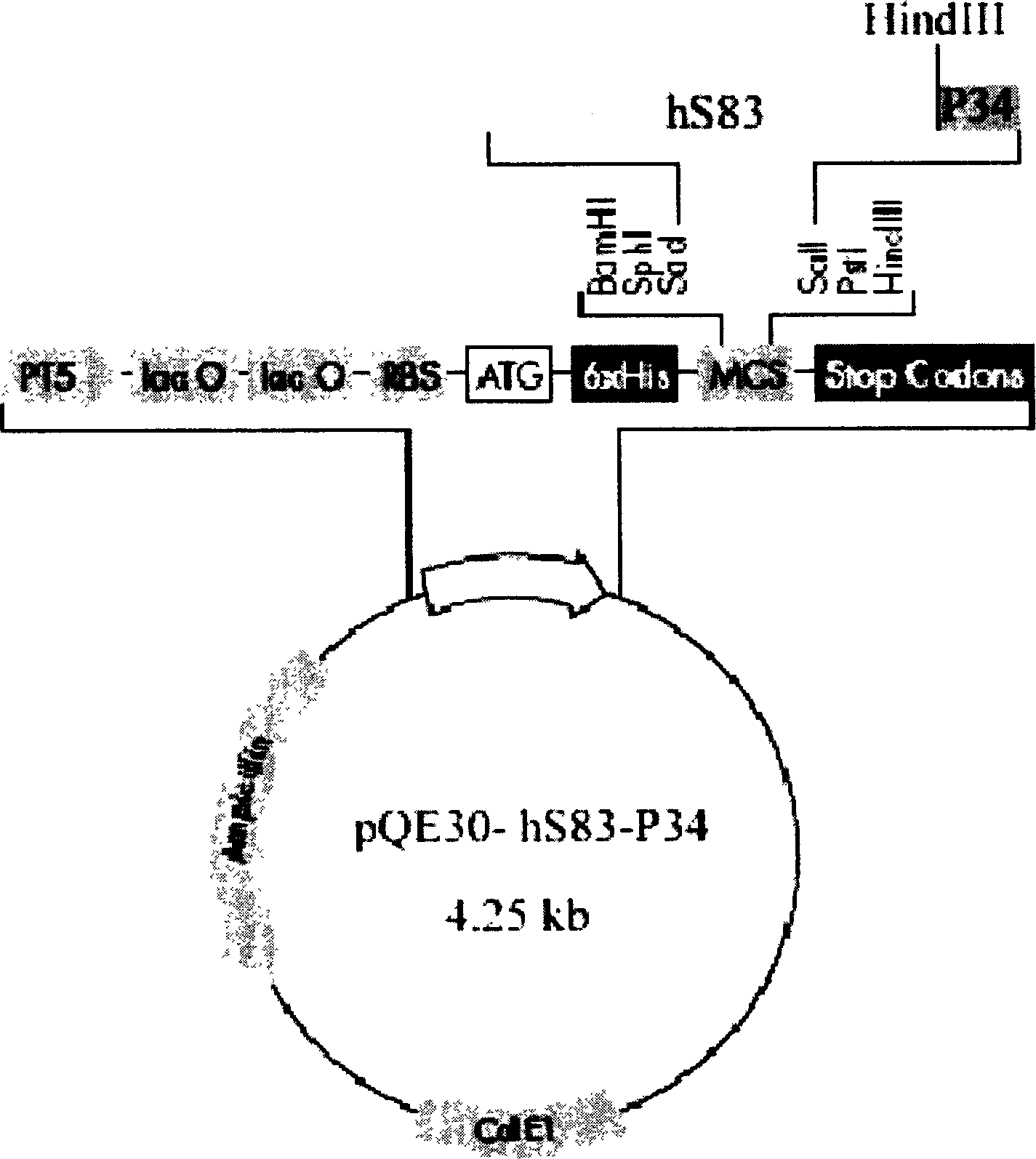
[0083] After the PCR product was purified by the Mini Gel Recovery Kit, it was digested with SacI / SalI, and then cloned into pBlueselect. SacI / SalI double enzyme digestion was used to identify, and the identified positive clones were re-sequenced for verification, and the plasmid with correct sequencing was named pBS-hS83. Restriction map see Figure 4 .
[0084] 2. Construction and induced expression of hS83 expression plasmid
[0...
Embodiment 3
[0094] The cell lines used in the following experiments were all purchased from the Cell Bank of the Chinese Academy of Sciences, and their product numbers and full names are as follows:
[0095] Raji: number TcHu44, tumor cell line of human Burkitt's lymphoma;
[0096] 6T-CEM: number TcHu2, human T cell leukemia cell line;
[0097] (The above two cell lines both constitutively express CTLA-4.)
[0098] ECV-304: numbered GNHu3, normal human umbilical vein endothelial cells.
[0099] 1. Cell killing specificity analysis
[0100] 1. Comparison of the killing effect of activated and non-activated T cells
[0101] Collect activated T cells (Ta) and non-activated T cells (To) by 1.5×10 5 Each well was inoculated in a 96-well plate, and then hS83-P34 was added to a final concentration of 1.5 μmol / L. After 24 hours, the amount of surviving cells was measured by MTS method at 490 nm, and hS83 was used as a control. Taking the cell survival rate of the PBS blank control group as 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com