Method of detecting and quantifying cytomegalovirus
A cytomegalovirus, specific sequence technology, applied in biochemical equipment and methods, microbial measurement/inspection, fermentation, etc., can solve the problem of amplification, poor detection efficiency, hindering oligonucleotide binding, primers and detection The probe does not know the problem, to achieve the effect of high sensitivity and rapid sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Preparation of intercalating fluorochrome-labeled oligonucleotide probes.
[0043] The phosphorus atom between the 4th base (G) and the 5th base (C) from the 5' end of the sequence shown in SEQ ID NO: 9 is labeled with the well-known pigment oxazolone as an intercalating fluorescent pigment, and Azoflavin-labeled oligonucleotide probe (SEQ ID NO: 9) (see Ishiguro, T (1996) Nucleic Acids Res, 24 (24) 4992-4997).
Embodiment 2
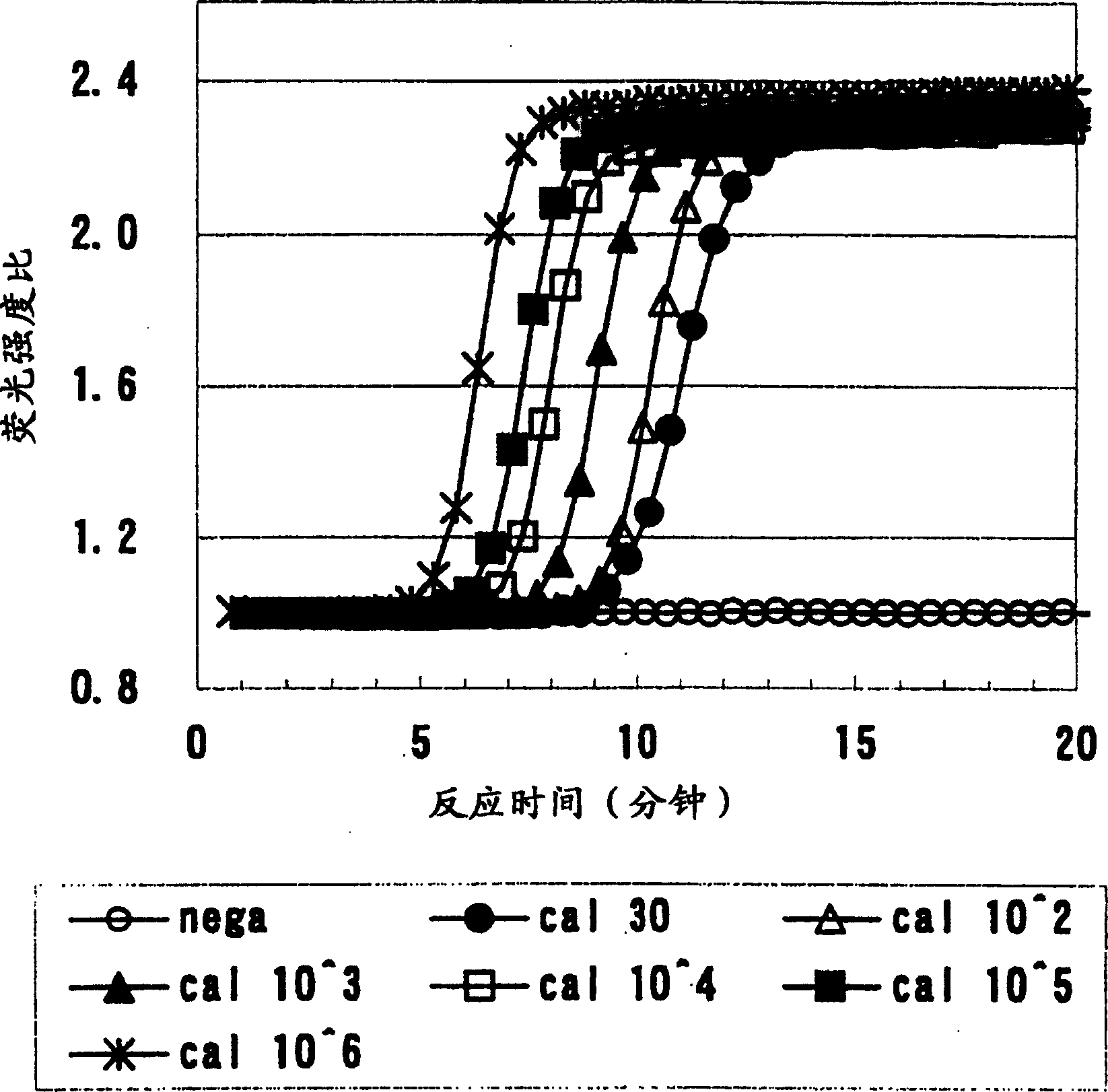
[0045] Using the combination of oligonucleotide primers invented by the present application, various initial copy numbers of β2.7 RNA are detected.
[0046] (1) Preparation of β2.7RNA
[0047] The so-called β2.7 RNA is a double-stranded DNA of 2154 bases in the base sequence of the β2.7 gene derived from CMV (accession number of the National Center for Biotechnology Information: 2471 bases from 184889 to 187359 of X17403) After cloning, synthesized and purified RNA is used as a template by in vitro transcription.
[0048] β2.7RNA (2154mer) was used as a sample, quantified according to the UV absorption at 260nm, and diluted with RNA diluent (10mM Tris-HCl (pH8.0), 0.1mM EDTA, 0.5U / μl ribonuclease inhibitor, 5.0mMDTT) into 1.0×10 6 copies / 5μl~30 copies / 5μl. Only RNA dilutions were used in the control experimental area (nega).
[0049] (2) 20.0 µl of a reaction solution having the following composition was dispensed into a 0.5 ml-capacity PCR tube (thin-walled reaction tube ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com