Application of leucocythemia related to gene ASH2L in leukemia molecule classification diagnosis
A technology for molecular typing and leukemia, applied in the fields of application, genetic engineering, plant gene improvement, etc., can solve the problems of uncertain expression characteristics of ASH2L, limitations of ASH2L in molecular typing diagnosis, unclear expression characteristics, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Primer name
Embodiment 2
[0027] Cloning of ASH2L, DPY-30-like and WDR5 genes
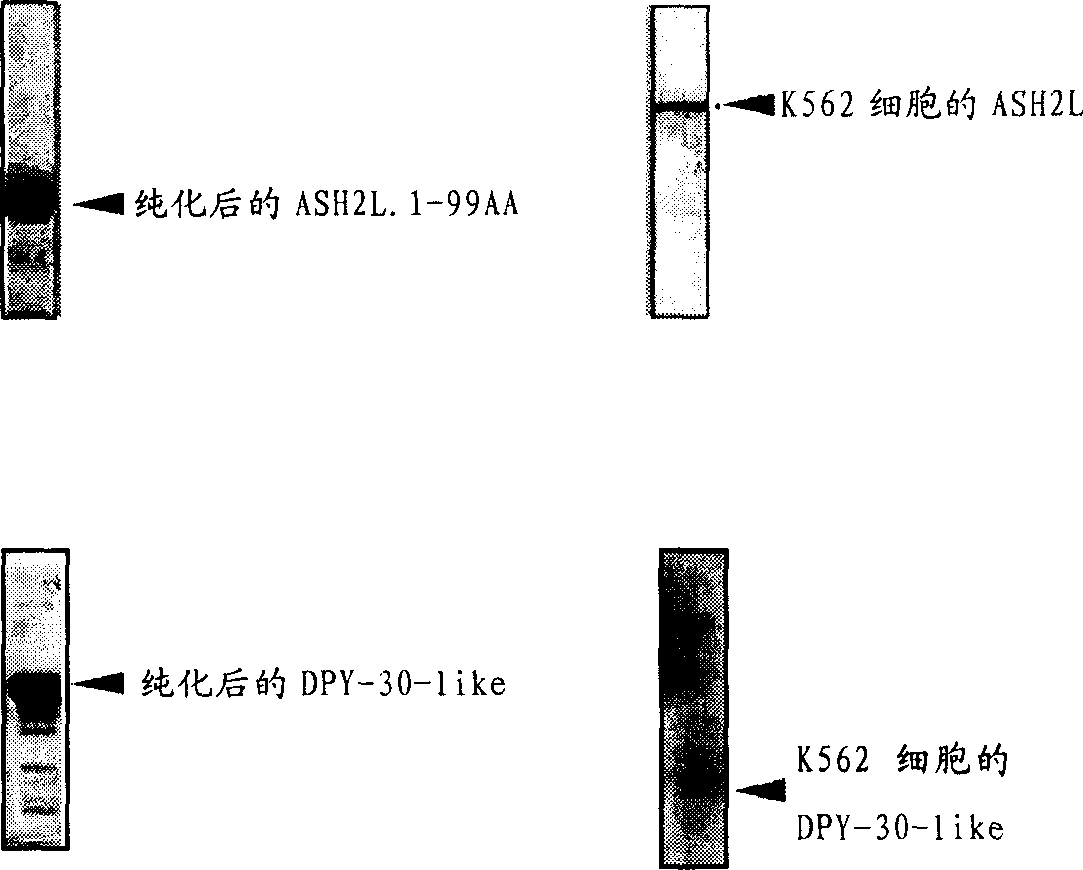
[0028] K562 cells were cultured on a 10cm dish with R1640 containing 10% FBS at 37 degrees, 5% CO 2 Cultured until the number of cells reached 2×10 7 Above, cells were harvested to extract total RNA and subjected to reverse transcription. Reverse transcription was carried out using a kit from PROMEGA, and the method is shown in the kit instructions. Using the cDNA as a template, three genes ASH2L, DPY-30-like and WDR5 were amplified. The primers used were ASH2L-1 / 2, DPY-1 / 2 and WDR5-1 / 2 (see Example 1 for the sequence). The conditions of the PCR reaction are: ASH2L, annealing temperature 64 degrees, extension 7 minutes; DPY-30-like, annealing temperature 60 degrees, extension 1 minute; WDR5 annealing temperature 60 degrees, extension 1.5 minutes; other conditions are the same Common PCR conditions, using Pfu enzyme.
[0029]After the PCR product was tailed with Taq enzyme at 72 degrees for 10 minutes, it was directly c...
Embodiment 3
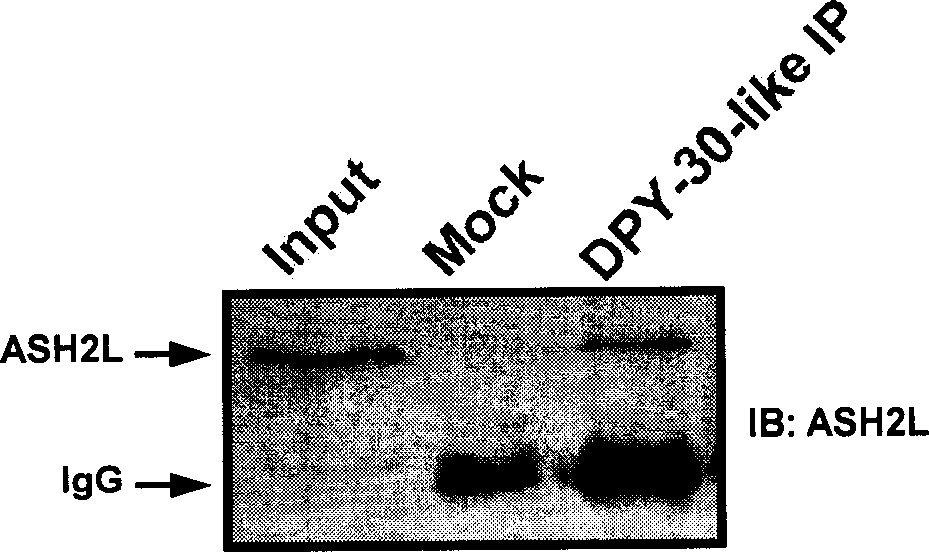
[0031] Preparation of anti-ASH2L, DPY-30-like specific antibodies:
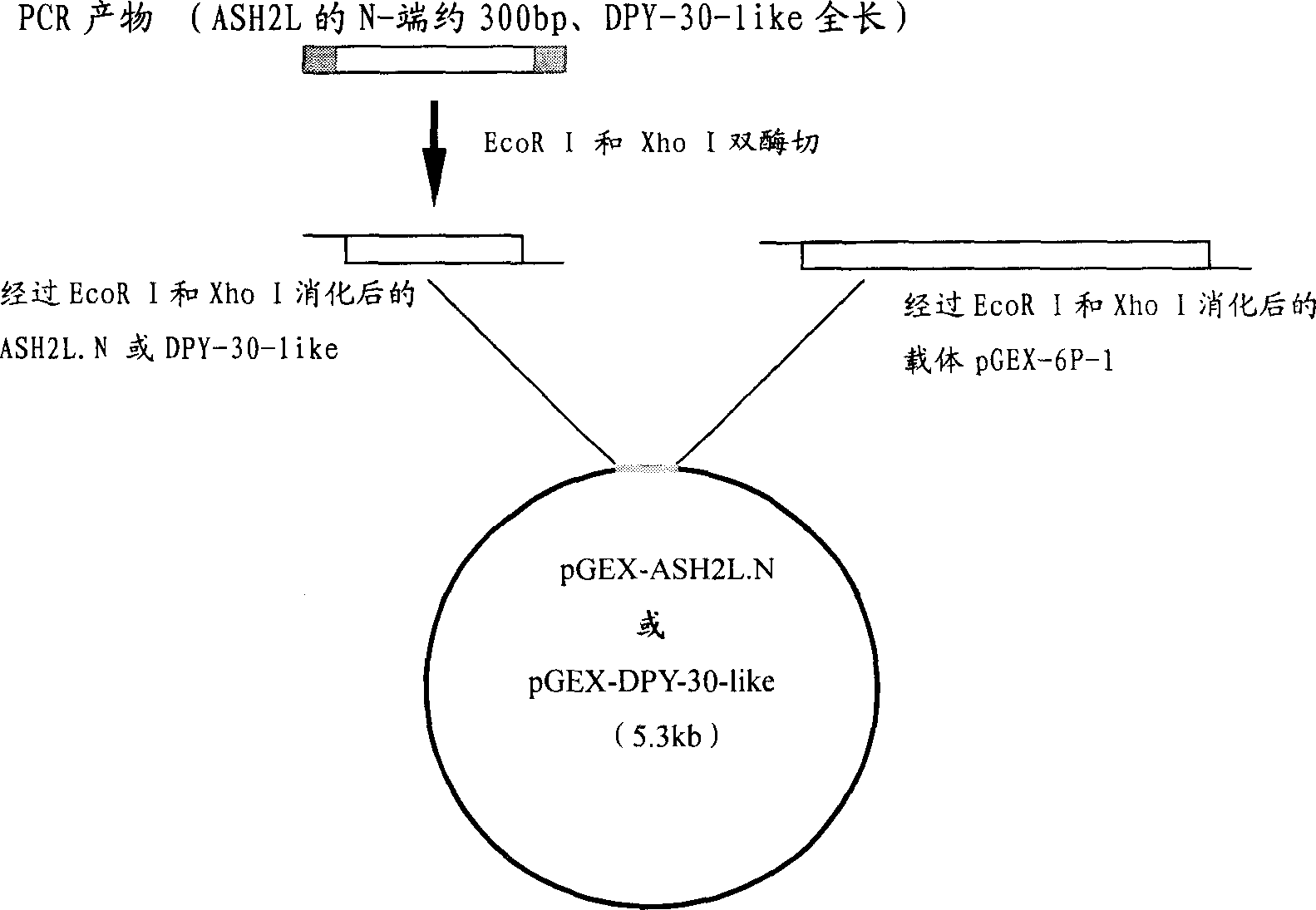
[0032] Antigen preparation: ASH2L and DPY-30-like cDNA template PCR amplification required fragments, primers are ASH2L-6P-UP / ASH2L-6P-DOWN and DPY-6P-UP / DPY-6P-DOWN two Right (see Example 1 for the sequence). The target fragment on ASH2L is the N-terminal 1-99aa coding region, and DPY-30-like is its full length. Both ends of the PCR fragment contain restriction enzyme sites EcoR I and Xho I, so these two enzymes were used to digest the PCR product and vector plasmid pGEX-6P-1. Then use T4 ligase to ligate the digested PCR fragment and the vector to form a recombinant plasmid (see figure 1 ).
[0033] PCR conditions: the annealing temperature is 65 degrees, the extension time is 1 minute, and other conditions are the same as ordinary PCR reactions. PCR reaction product 50ul (approximately 5ug) and vector plasmid (pGEX-6P-1) 6-10ug were digested with EcoR I and Xho I respectively, and DNA fragments were re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com