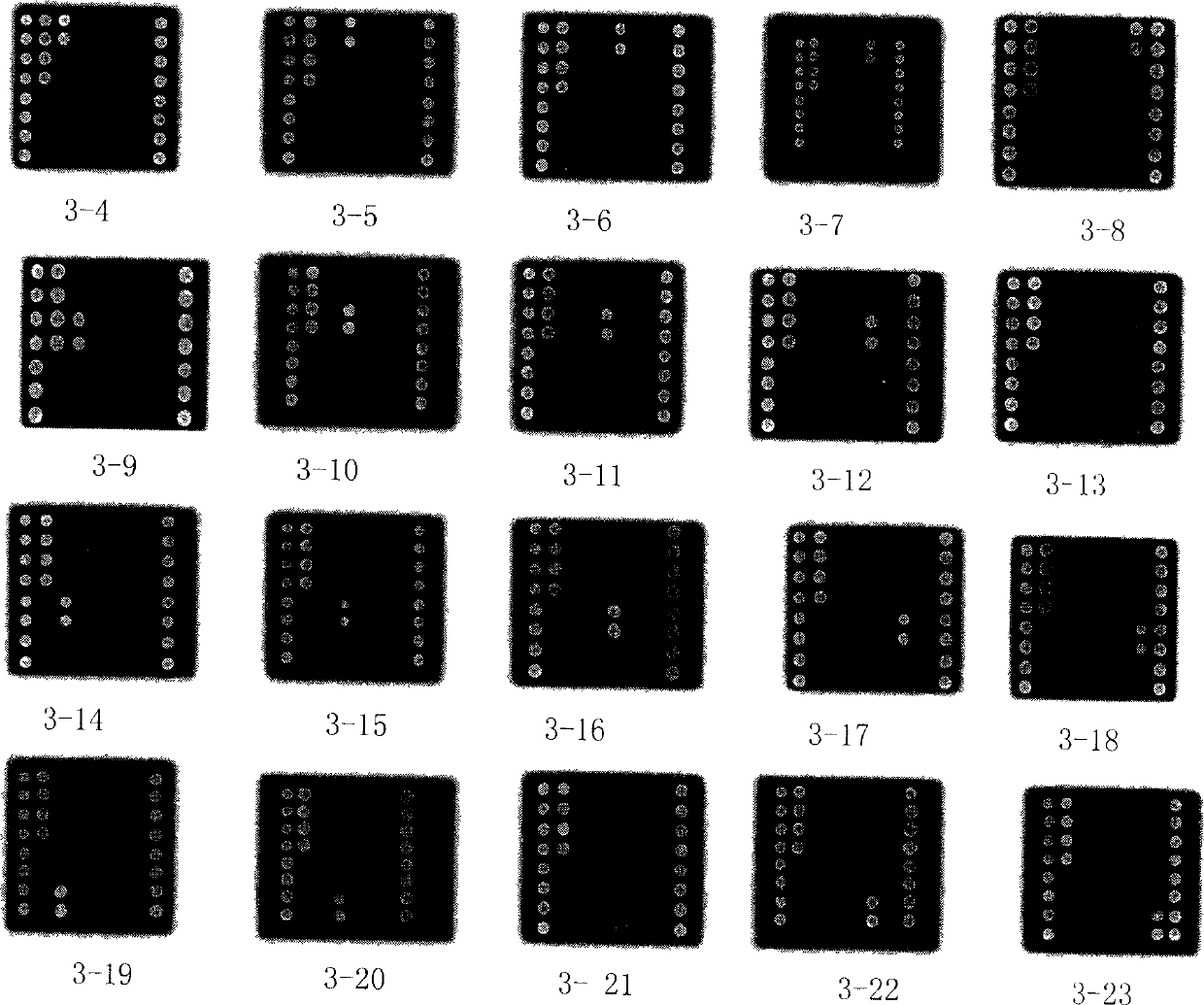
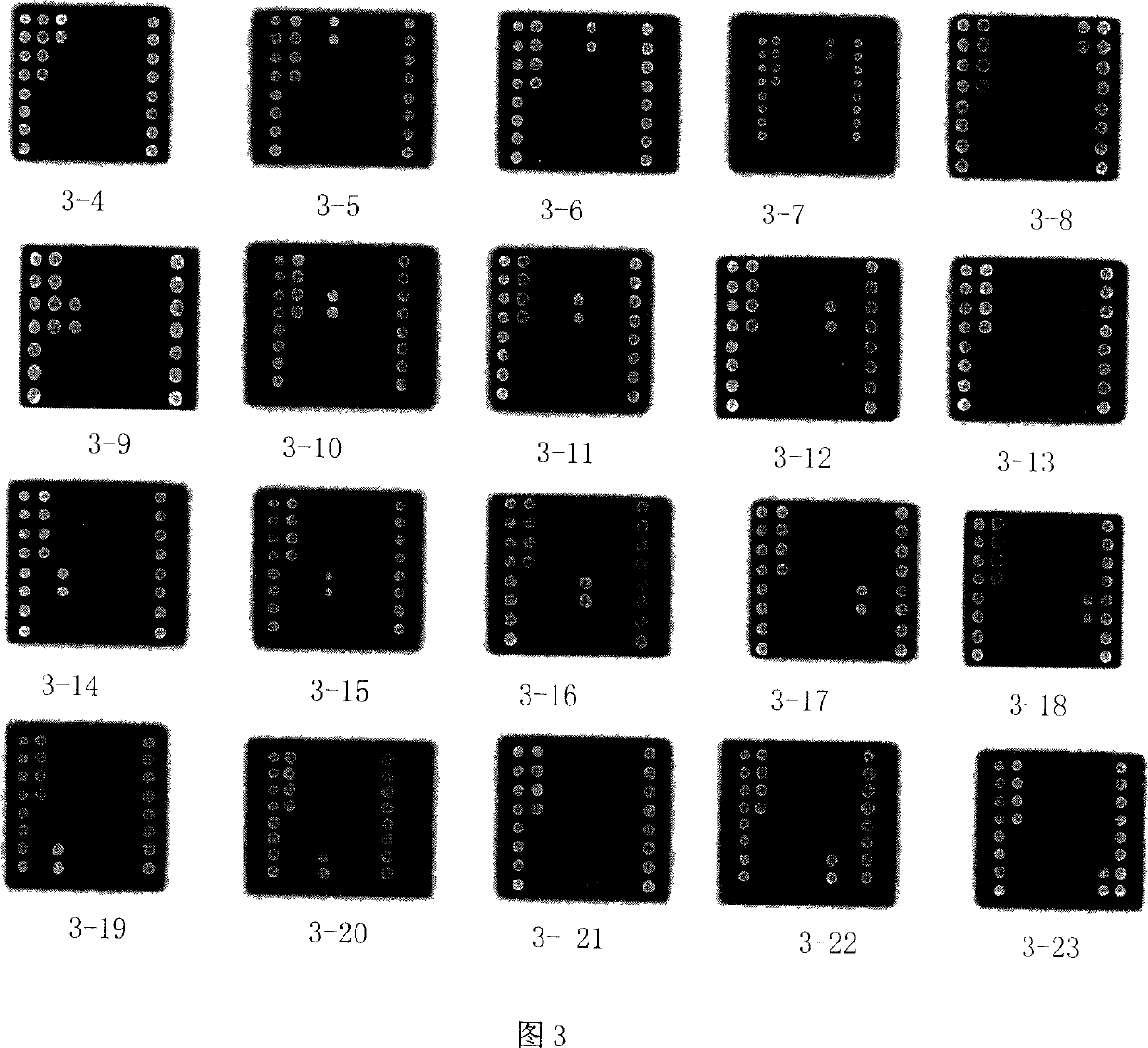
Biochip for detecting pathogenesis fungus
A technology of biochips and pathogenic fungi, applied in the determination/testing of microorganisms, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0079] Establishment of fungal culture conditions
[0080] 1. Yeast (Candida and Cryptococcus)
[0081] Apply Sandcastle solid or liquid medium and culture at 30°C for 2 days
[0082] Recipe: Glucose 40g
[0083] Peptone 10g
[0084] Agar 20g
[0085] Distilled water 1000ml
[0086] Mix the above ingredients, heat until completely dissolved, adjust the pH to about 5.6, and steam sterilize at 115°C for 30 minutes for later use
[0087] Note: liquid medium without agar
[0088] 2. Filamentous fungi and superficial dermatophytes
[0089] Use potato solid or liquid medium for cultivation at 30°C for 5-7 days
[0090] Recipe: Boil 200 peeled and chopped potatoes with 700ml of water for 20 minutes, filter with gauze, and keep the filtrate
[0091] Add glucose 20g
[0092] Agar 15g
[0093] Add water to 1000ml,
Embodiment 2
[0095] Establishment of DNA Extraction Method from Fungi
[0096] Extraction of Fungal DNA Using Benzyl Chloride
[0097] 1. Principle
[0098]
[0099] ROH stands for chitin in the cell wall, above for PhcH 2 cL in the reaction formula of various polysaccharides
[0100] 2. Method
[0101] (1) Scrape 0.1-0.3g of colonies with a volume of about 100ul into a 1.5ml centrifuge tube
[0102] (2) Add 600μl DNA extraction solution, shake and mix
[0103] (3) Add 10% SDS 50ul shake and mix
[0104] (4) Add 300ul benzyl chloride and shake to mix
[0105] (5) 50-55 ℃ water bath for 1 hour and mix every 10 minutes
[0106] (6) Add 60ul of 3M sodium acetate to each tube and bathe in ice for 20 minutes
[0107] (7) Centrifuge at 4°C, 13000rpm for 10 minutes
[0108] (8) Take the supernatant, add an equal volume of phenol chloroform, and centrifuge at 13,000 rpm for 10 minutes at 4°C
[0109] (9) Take the supernatant, add 1 / 10 sodium acetate and an equal volume of isopropan...
Embodiment 3
[0116] Target gene selection
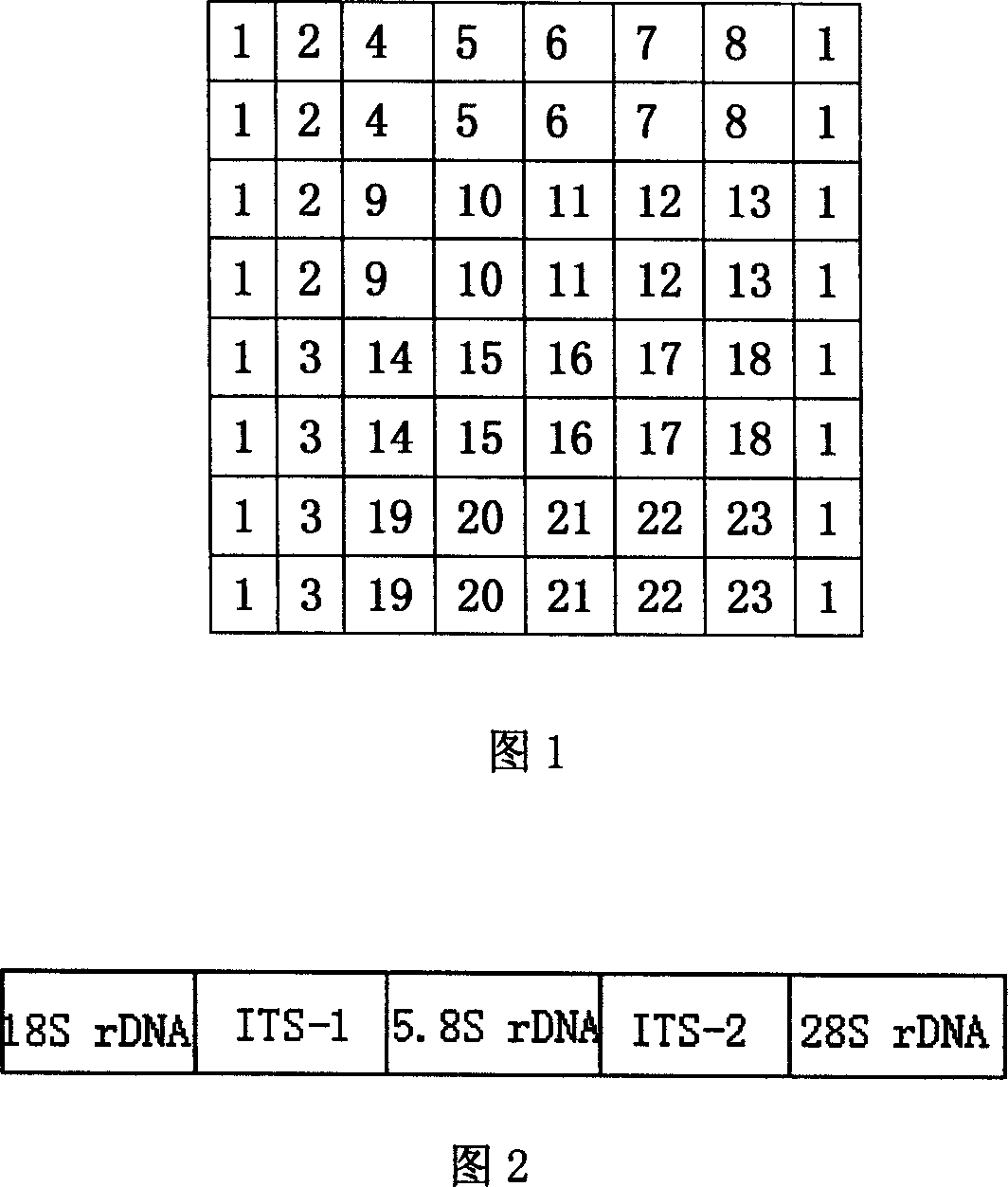
[0117] Whether in fungi or bacteria, cellular ribosomal ribonucleic acid (rRNA) genes are commonly used targets for molecular analysis. The ribosomal ribonucleic acid (rRNA) gene and its adjacent spacer are collectively referred to as rDNA. Fungal cell rDNA has dozens or even hundreds of copies, and the fungal ribosomal DNA (rRNA gene) operator includes three functional genes, namely 18S rDNA, 5.8S rDNA, and 28S rDNA, which are connected head to tail on the chromosome, arranged in series, and mutually Between the internal transcribed spacer (internal transcribed spacer) division. Including the internal transcribed spacer-1 (ITS-1) between 18S rDNA and 5.8S rDNA, and the internal transcribed spacer-1 (ITS-2) between 5.8S rDNA and 28S rDNA. (See Figure 2)
[0118] The nuclear rDNA of eukaryotes is a multi-gene cluster, arranged in tandem repeat units, and a typical rDNA repeat unit is about 8-12kb. They are located in nucleolar organizer region...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com