Mammalian cell tandem affinity purification vector and method for purifying protein complex thereof
A carrier and affinity technology, applied in the field of protein separation and purification, to reduce the influence of function or stability, and the effect of reducing the molecular weight of tags
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1 Construction of tandem affinity purification vector p2FCBP
[0030] The tandem affinity purification vector is developed on the basis of the retrovirus vector MSCVpac. The TAP tag sequence composed of 2×Flag, TEV enzyme digestion sequence and CBP sequence plus the linker sequence of HindIII and BamH I restriction endonucleases, and the vector that was double-digested with restriction endonucleases HindIII and BamH I MSCVpac is connected.
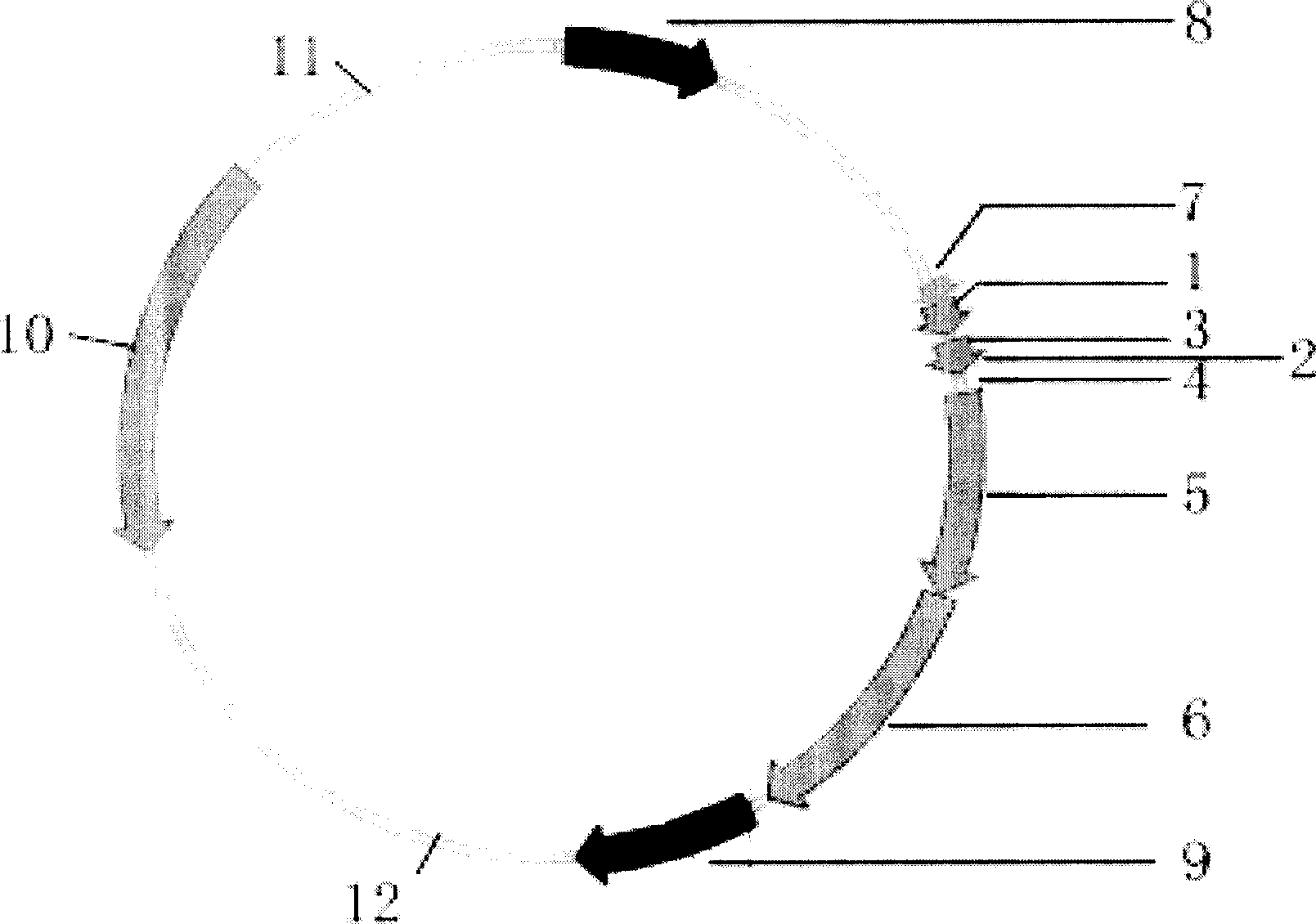
[0031] Prepare tandem affinity purification vectors such as figure 1 As shown, named p2FCBP. The p2FCBP vector has multiple cloning sites such as BamH I, BstX I, EcoRV, and Xho I for convenient insertion of foreign genes, and two identical 8-peptide Flag tags (DFlag) and one Calmodulin-binding peptide (CBP) sequence, there is a TEV enzyme cleavage site between the Flag tag and the CBP sequence. There is also a screening marker encoding ampicillin (Amp) and puromycin N-acetyl transferase (puromycin N-acetyl transferase) o...
Embodiment 2
[0032] Example 2 Application of the p2FCBP vector to identify the interacting protein of the target protein 14-3-3ε in QGY7703 cells
[0033] (1) Cell culture and collection
[0034] QGY7703 was cultured in culture medium 1640 containing 10% fetal calf serum (FCS) at 37°C and 5% CO 2 , passaging every 2-3 days. When the cells grow to 80-100%, treat with 0.25% trypsin / 0.02% EDTA digestion solution for a few minutes to detach the cells, add pre-cooled PBS to suspend, centrifuge to wash off the culture medium, and repeat with PBS again.
[0035] (2) RT-PCR
[0036] 10 will be collected 6 Extract total RNA from QGY7703 cells and use 50 μl RNase-free ddH 2 O dissolved. The sense primer (5'-TATGGATCCGATGATCGAGAGGATCTGGTGTAC-3') for PCR amplification of 14-3-3ε; the antisense primer (5'-ATTCTCGAGTCACTGATTTTCGTCTTCCACGTC-3'). RT-PCR conditions are: 2 μl of total RNA as template, 1 μl of 20 μmol / L primers, 5 μl of 10 mmol / L dNTP, 25 mmol / L MgCl 2 10μl, 5μl of 10×PCR buffer, 1μl ...
Embodiment 3
[0047] Example 3 Application of the p2FCBP vector to identify the interacting protein of the target protein 14-3-3ε in MHCC97H cells
[0048] The steps are the same as in Example 2, except that the p2FCBP recombinant vector inserted with 14-3-3ε cDNA is transferred into MHCC97H cells for fusion protein expression. Final protein profiling revealed that the components in the 14-3-3ε complex were 14-3-3β and keratin CK 1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com