Wheat gibberella resistance major effect QTL linked molecular label and its application
A wheat scab and molecular marker technology, applied in the fields of application, botanical equipment and methods, plant gene improvement, etc., can solve the problems that there is no molecular marker yet, it is obviously better than Sumai 3, and has been used by breeders, etc. , to achieve the effect of improving accuracy and breeding efficiency and reducing workload
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach
[0018] Specific embodiments: materials and methods: 1. Ning 894037 / Alondra recombinant inbred line population construction and inoculation anti-red identification:
[0019] F 1 bastard, F 1 F 2 Offspring, F 2 The 219 individual plants randomly select a seed to plant, adopt the method of single-seed transmission, and propagate to F 8 generation, resulting in 219 families.
[0020] The identification of head blight resistance adopts the method of single flower dripping. The concentration of the conidia suspension of Fusarium graminearum is 50000 / ml. Select the ears that have just bloomed, and add 10ul of the spore suspension to the middle floret, and mist the greenhouse to keep it moist. For 3 days, in the field, bagging or artificial fogging was used to keep moisture. Each line was inoculated with 10 to 12 ears, and the rate of diseased spikelets was investigated 21 days after inoculation. Diseased spikelet rate=(number of sick spikelets / total spikelet number)×100%
[002...
Embodiment 1
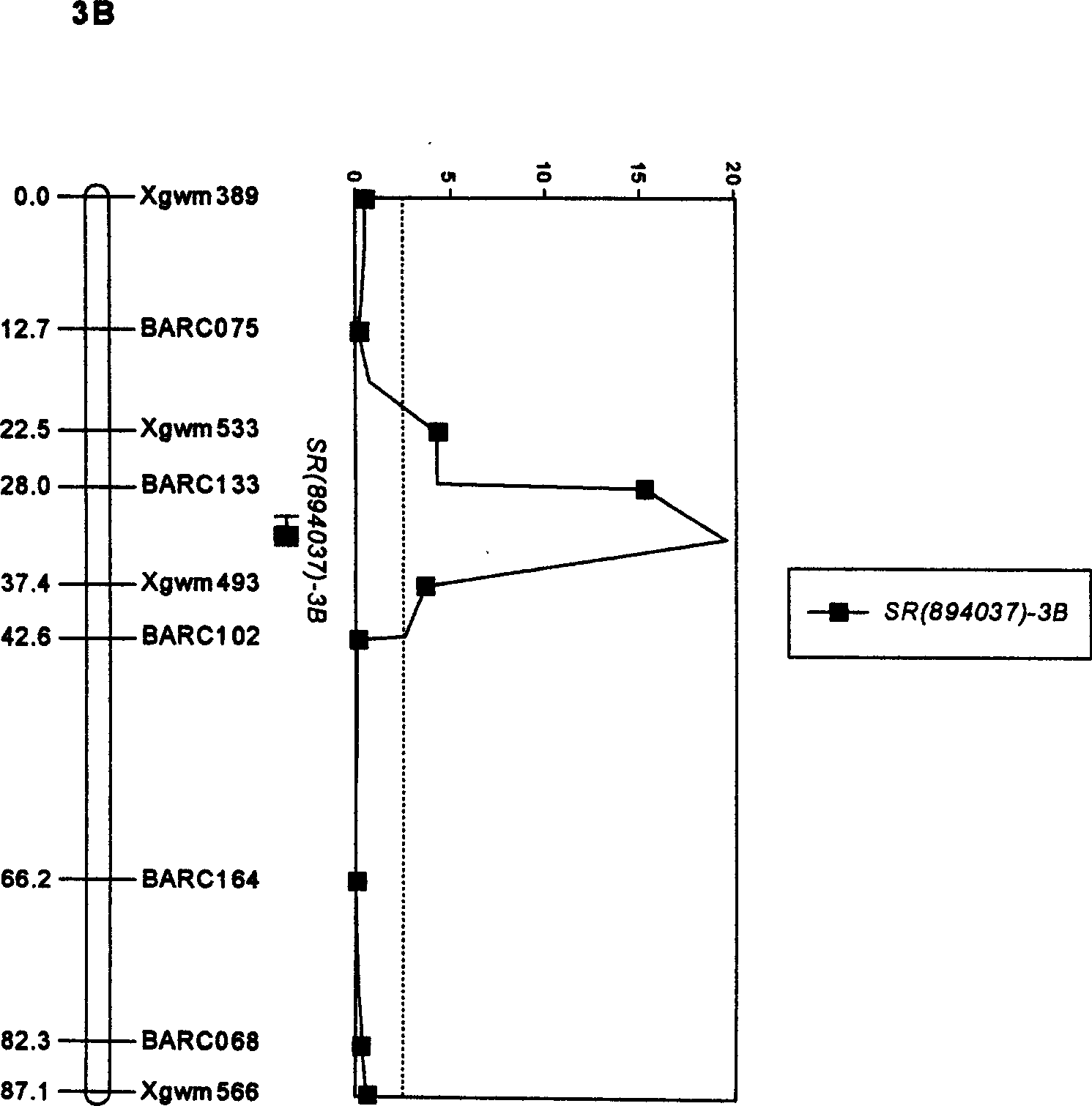
[0024] Molecular markers linked to major QTLs for wheat head blight resistance were obtained by the following methods:
[0025] a) The wheat scab-resistant variety Ning 894037 (♀) was crossed with the wheat scab-susceptible variety Alondra (♂) to obtain the hybrid F 1 .
[0026] b) by Hybrid F 1 Self-pollination to obtain F 2 Multiple offspring, get F by single grain pass (SSD) method 8 generation of recombinant inbred lines.
[0027] c) Molecular marker analysis is performed on each strain of the recombinant inbred line, and the genotype of each strain of the recombinant inbred line is described; the specific method: separate the genomic DNA of each strain of the recombinant inbred line, and use SSR Primers carry out PCR amplification, amplified products are separated by electrophoresis on polyacrylamide gel of 6% (containing 5.7 gram acrylamide and 0.3 gram methylene acrylamide in 100ml polyacrylamide gel solution), after silver staining, obtain each strain genotype.
...
Embodiment 2
[0033] Using the screened molecular markers linked to the main QTL for scab resistance in Ning 894037, a preliminary test was carried out on the advanced breeding materials to test the practical value of this method in molecular marker-assisted selection for wheat scab resistance. Select 6 high-generation breeding materials or wheat varieties with Ning 894037 and its derivative varieties (lines) as male or female parents, isolate their genomic DNA, and then use SSR primers Xgwm533, BARC075 and BARC133 to perform PCR reactions on these DNA, electrophoresis Finally, determine whether there is a corresponding marker according to the size or presence of the DNA band. If there are 2-3 markers at the same time, it means that the resistance of the variety or strain to scab has reached the level of "resistance", and if it does not exist, it is susceptible , at the same time, it was verified by the results of inoculation identification and molecular marker detection of scab resistance. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com