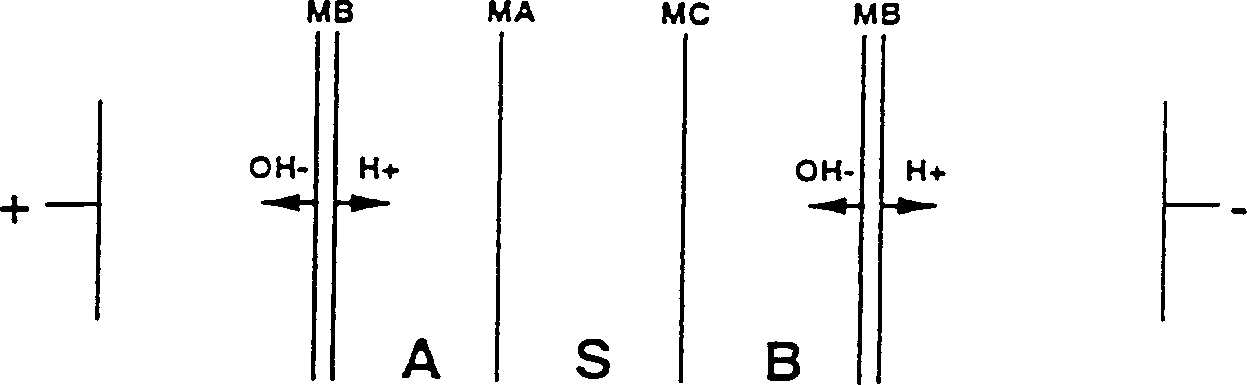
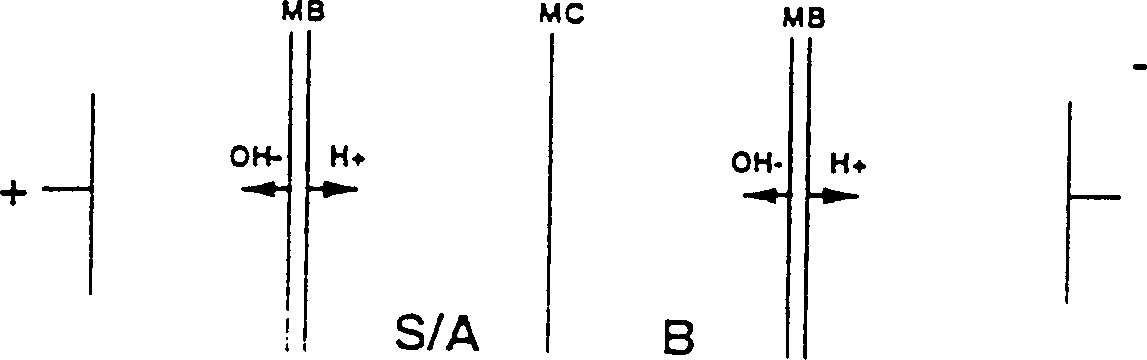
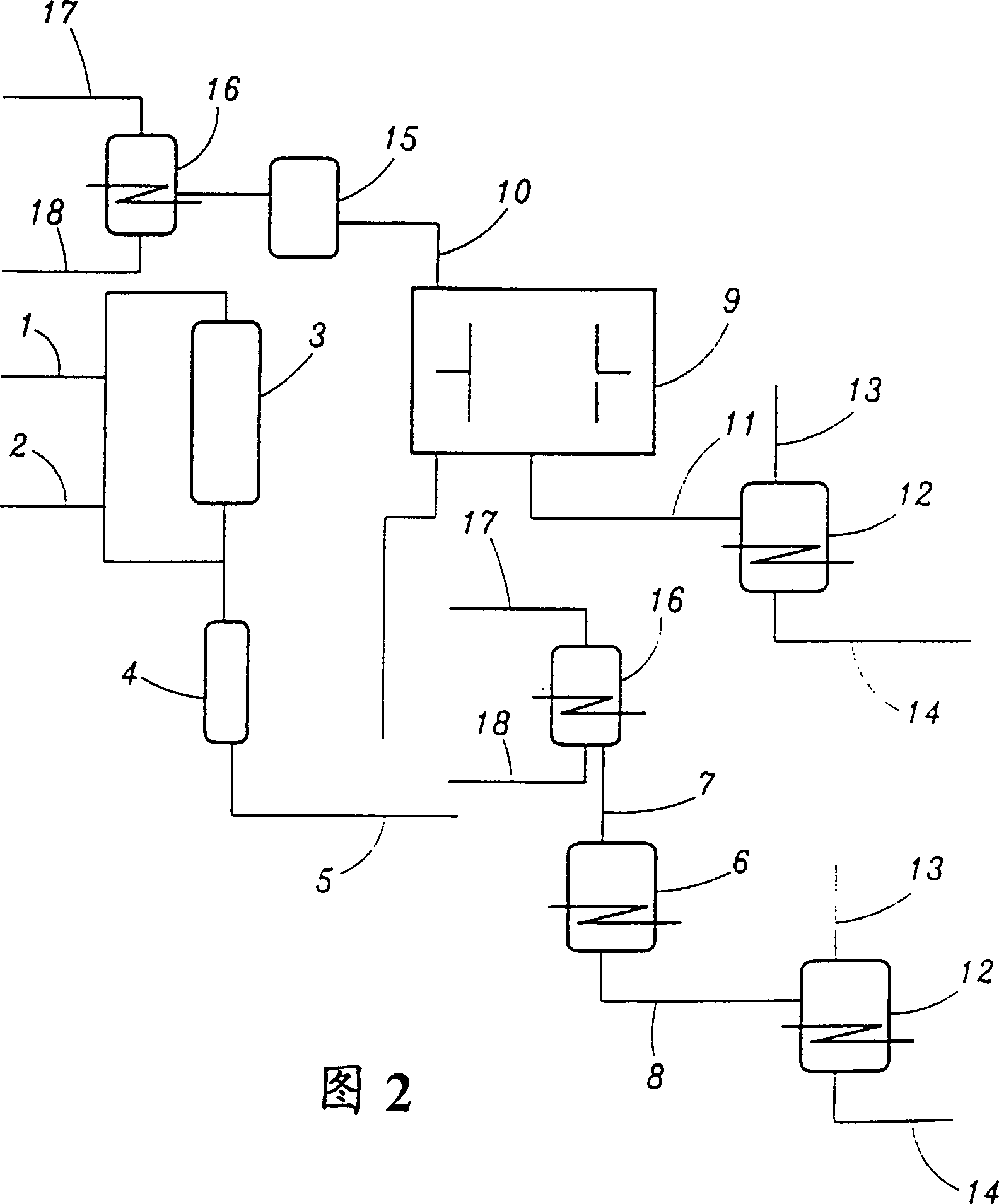
Method for preparing 2-hydroxy 4-methylthio butyric acid using nitrilase
A technology of methylthiobutyric acid and methylthiobutyric acid ammonium salt is applied in the field of preparing 2-hydroxy-4-methylthiobutyric acid with nitrilase, which can solve the problem of insufficient microbial activity of nitrilase and the unusable issues such as industrial scale
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0120] Example 1: Purification and N-terminal sequencing of nitrilase
[0121] The nitrilase was purified in four steps. A summary of the purification is shown in Table 1 below.
[0122] Alcaligenes faecalis cells were grown for 24 hours at 30°C in minimal medium containing benzonitrile (0.5 g / l). After the culture was centrifuged, the pellet was resuspended in TG buffer (25 mM Tris-HCl, 10% (w / v) glycerol, pH 7.5). The cell suspension was sonicated and then centrifuged to obtain a crude extract. The crude extract was then treated with ammonium sulfate until 30% saturation. The resulting pellet was resuspended in TG buffer and then dialyzed overnight against 2 liters of the same buffer. The resulting solution was then loaded onto a Q Sepharose Fast Flow HR 26 / 10 anion exchange column pre-equilibrated with TG buffer. The active ingredient was then eluted with a gradient of 0-1M NaCl. The active fraction was loaded onto a Mono Q HR5 / 5 anion exchange column pre-equili...
Embodiment 2
[0126] Embodiment 2: Cloning of Alcaligenes faecalis ATCC8750 nitrilase
[0127] The N-terminal sequence given in Example 1 and the N-terminal sequence of Alcaligenes faecalis JM3 nitrilase (Kobayashi et al., 1993, Proc. Natl. Acad. Sci. USA 90: 247-251 ) were completely identical, while the N-terminus of bacterial nitrilases had 35-57% identity over 14 residues. The inventors hypothesized that the nitrilase of the present invention purified from the ATCC8750 strain was identical to the enzyme described by Kobayashi et al. (cited above). Therefore, the cloning strategy is to carry out PCR reaction on the DNA genome of the ATCC8750 strain with two nucleotide probes determined according to the sequence given by Kobayashi et al. (cited above) to amplify the nitrilase gene.
[0128] Two probes were synthesized, one hybridizing to the 5' portion and the other hybridizing to the 3' portion of the sequence given by Kobayashi et al. (cited above):
[0129] 5' portion (PCRAF1...
Embodiment 3
[0132] Embodiment 3: the sequencing of the 1130pb fragment containing nitrilase activity polypeptide coding DNA
[0133] The insert cloned in plasmid pRPA-BCAT3 has been sequenced by Genome Express S.A. (Grenoble, France) using laboratory prepared DNA preparations (kit Wizzard Midi-prep, Promega). The sequencing strategy for this fragment is shown in Figure 4 , and implemented by conventional methods known to those skilled in the art. Ten internal nucleotide primers (in Figure 4 Winning designations 623-629 and 681-682). The full-length sequence was completed with universal primers "Reverse" and "M13 Forward". Each region is read at least once on each DNA.
[0134] The resulting sequence differs from the published sequence in two places: one in the putative structure serving as a transcription terminator, and the other in the gene for the nitrilase enzyme (called nitB), leading to Asn 279 → Replacement of Asp. Thus on the plasmid pRPA-BCAT1,2,4,5 with two spec...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle size | aaaaa | aaaaa |
| particle size | aaaaa | aaaaa |
| particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com