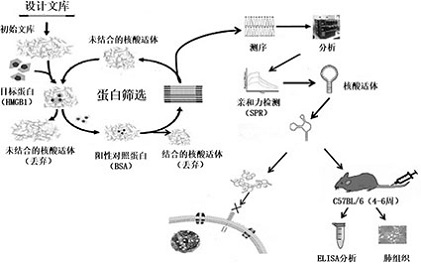
Nucleic acid aptamers targeting and antagonizing HMGB1 molecules
A nucleic acid aptamer and antagonism technology, which is applied in the field of HMGB1 nucleic acid aptamer antagonists, targeting and antagonizing HMGB1 molecular nucleic acid aptamers, can solve the problems of low immunogenicity and achieve low immunogenicity, small molecular weight, good affinity effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0096] Preparation of nucleic acid aptamers
[0097] 1. Construction of screening library
[0098] The ssDNA library used in this study has a full length of 70 nt, consisting of 20 nt primer fragments fixed at both ends and a random fragment of 30 nt in the middle; the 5'-modified FITC fluorophore of the forward primer is fixed to the 5' end of the initial library The sequence is the same; the 5' end of the reverse primer is labeled with Biotin, and the sequence is reverse complementary to the fixed sequence at the 3' end of the original library;
[0099] The sequence of the screening library targeting HMGB1 is:
[0100] SEQ ID NO. 3: AAGGAGCAGCGTGGAGGATA (N) 30 TTAGGGTGTGTCGTCGTGGT.
[0101] The primers used to construct the library are:
[0102] Forward primer: SEQ ID NO. 4: AAGGAGCAGCGTGGAGGATA;
[0103] Reverse primer: SEQ ID NO. 5: ACCACGACGACACACCCTAA.
[0104] 2. Conjugation of protein and magnetic beads
[0105] Take the carboxyl magnetic beads (MB), wash them w...
Embodiment 2
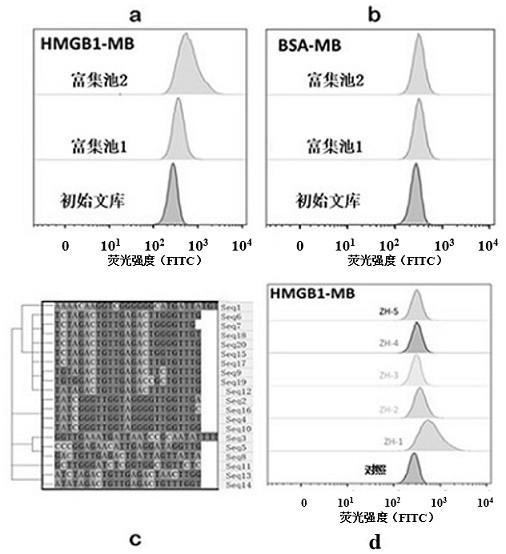
[0115] Validation of nucleic acid aptamer binding to HMGB1 protein
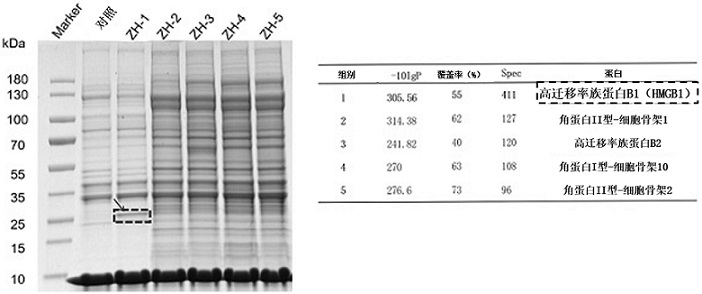
[0116] The protein of SKOV3 cells was extracted and divided into 6 equal parts. The biotin-labeled control sequence and candidate chain were added to the protein, mixed well, incubated at 4°C for 45 min, and then 50 μl of streptavidin-modified magnetic beads were added to it. , after mixing, continue to incubate at 4 °C for 1 h, after the incubation, remove the supernatant for washing 3 times, then add 2× protein loading buffer to the tube, mix well, denature at 100 °C for 10 min, and separate the protein , and finally take the supernatant and load the sample, and use SDS-PAGE for electrophoresis. After the electrophoresis is completed, the gel is stained with Coomassie brilliant blue to identify the different bands.
[0117] Test results such as image 3 As shown, according to the test results, it can be seen that the nucleic acid aptamer ZH-1 can specifically recognize and bind the HMGB1 protein in the cel...
Embodiment 3
[0119] Optimization of nucleic acid aptamer ZH-1 sequence
[0120] Through the sequence truncation strategy, ZH-1 was truncated into 6 optimized sequences, and the affinity of the 6 sequences was determined by WesternBlot, and the internal reference was tublin; the detection results are as follows Figure 4 shown, by Figure 4 It can be seen that ZH-1a (SEQ ID NO. 2) has the strongest affinity, so ZH-1a is determined to be the optimal nucleic acid aptamer for subsequent experiments.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com