Nucleic acid aptamer for recognizing extracellular vesicles of ovarian cancer drug-resistant patient and application of nucleic acid aptamer
A nucleic acid aptamer, drug-resistant cell technology, applied in the biological field, can solve the problems of low toxicity and immunogenicity, and cannot be used to detect drug resistance in ovarian cancer patients, and achieves strong affinity and specificity, low cost, good recognition effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
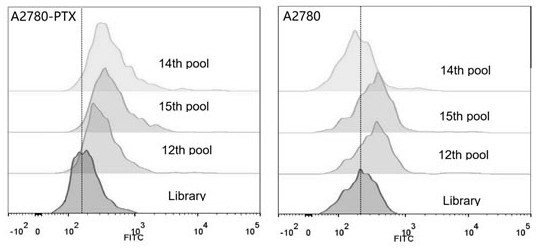
Cell-SELEX screening process
(1) Design of nucleic acid library and primers:
Random nucleic acid library:
5'-AAGGAGCAGCGTGGAGGATA(N)45TTAGGGTGTGTCGTCGTGGT-3';
Upstream primer: 5'-fluorescein isothiocyanate-AAGGAGCAGCGTGGAGGATA-3';
Downstream primer: 5'-biotin-ACCACGACGACACACCCTAA-3';
(2) Positive sieve:
The above random nucleic acid library was placed in a centrifuge at 10,000 rpm, centrifuged for 10 min, fully dissolved in DPBS, denatured at 95°C for 5 min, and quickly placed in ice for 10 min.
[0037] ②Incubate:
The treated random library was incubated with the cultured and pretreated ovarian cancer paclitaxel-resistant cell line A2780 / PTX at 37°C for 60 min.
[0038] ③Separation:
After the incubation, remove the solution in the incubation dish, wash the cells in the incubation dish with washing buffer (PBS, containing 0.45% glucose, 5mM magnesium chloride); scrape the cells in the incubation dish with 600 μL of pre-cooled sterile water...
Embodiment 2
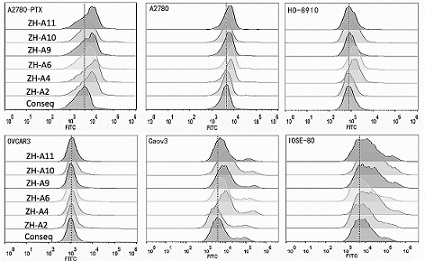
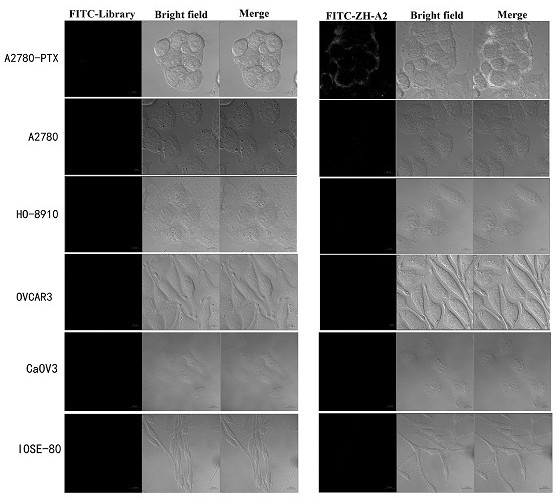
Determination of optimal nucleic acid aptamers
According to the nucleic acid library screened in Example 1, 6 candidate nucleic acid sequences were selected through computer analysis and comparison, and were named ZH-A2, ZH-A4, ZH-A6, ZH-A9, ZH-A10, ZH- A11 and determine the optimal nucleic acid aptamer.
[0046] Specific steps:
(1) Streaming inspection
Adherent state A2780 / PTX cells were digested from culture dishes with 0.2% EDTA, cells were collected into centrifuge tubes, and washed twice with DPBS; were added to A2780 / PTX cells soaked in binding buffer (D-PBS, containing 0.45% glucose, 5mM magnesium chloride, 100mg / L tRNA, 1g / L BSA); then placed in a shaker at 37°C and incubated for 45min; the incubation was completed After that, it was centrifuged and washed twice with washing buffer (PBS, containing 0.45% glucose, 5mM magnesium chloride), and finally the fluorescence was detected by flow cytometry;
The same method was used to detect the binding of candidate seque...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com