Method for simultaneously enhancing expression quantity and solubility of target protein in prokaryotic system
A protein expression and solubility technology, applied in the biological field, can solve the problems of unpredictable target protein interaction and reduced work efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Embodiment 1: Construction of fusion protein VeGFP
[0035] This example exemplifies a fusion protein VeGFP spliced with the N-terminal signal peptide of the naturally occurring protein Vip3Aa (SEQ ID NO: 1) and eGFP protein.
[0036] The characteristics of the VeGFP protein are: the N-terminal signal peptide sequence of Vip3Aa is located at the N-terminal of the eGFP protein; there are no other redundant amino acids between the N-terminal signal peptide of Vip3Aa (SEQ ID NO: 1) and the amino acid sequence of the eGFP protein. The polynucleotide sequence encoding the VeGFP protein can be obtained from at least three approaches: 1) by obtaining the polynucleotide sequence of the N-terminal signal peptide encoding Vip3Aa and the amino acid sequence of the eGFP protein with a polymerase chain reaction (PCR reaction), and then The two are spliced together by overlapping PCR (Horton, R.M.et al.BioTechniques, 1990) to form a complete fusion gene; 2) by combining the N-ter...
Embodiment 2
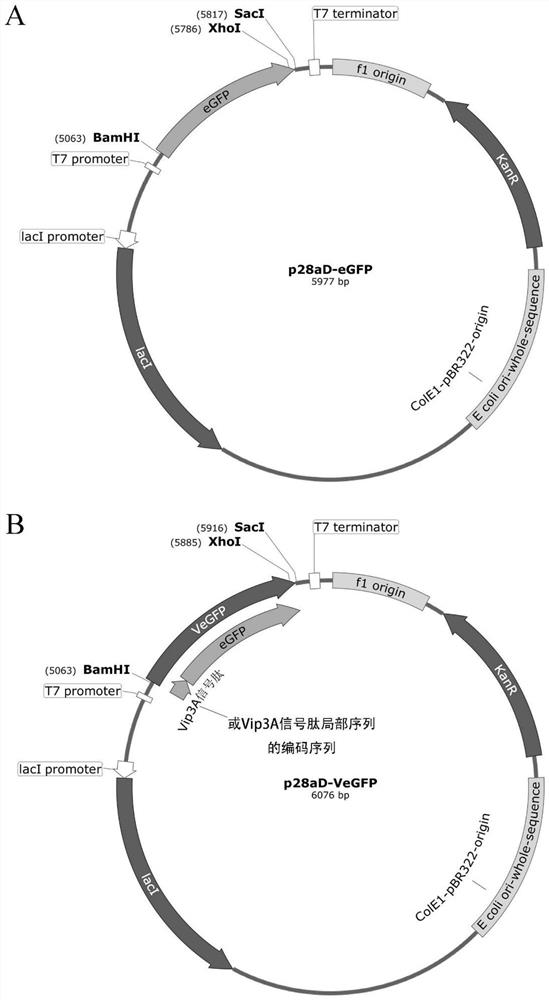
[0037] Example 2: Construction of the prokaryotic cell expression vector p28aD-VeGFP of the fusion protein VeGFP
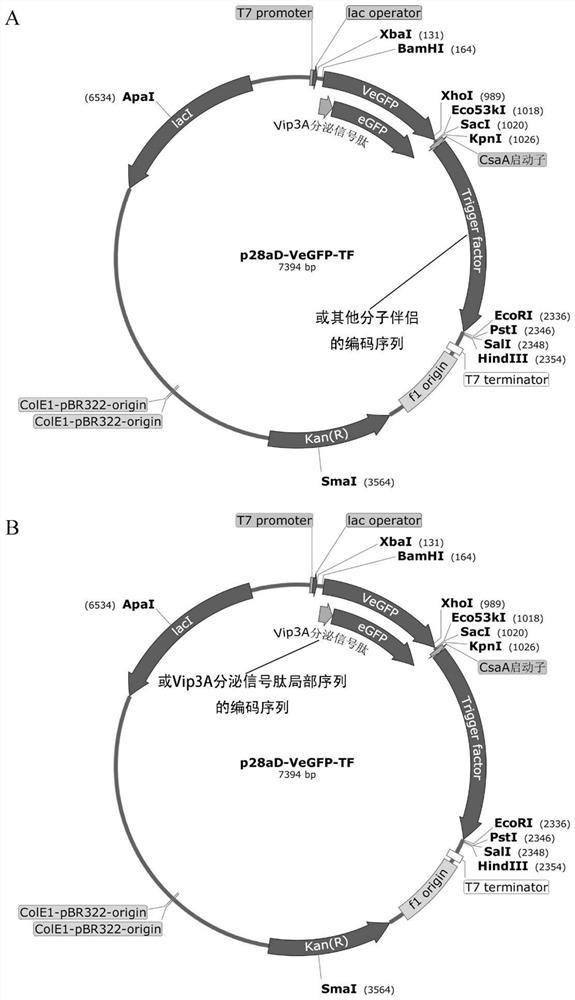
[0038] This example illustrates a prokaryotic expression vector for the fusion protein described in Example 1 herein. First, the two ends of the VeGFP gene were added to BamHI (5') and SacI (3') sites respectively. Then the gene was inserted into the corresponding site of pET28aDel vector (Gao et al.2011) to form p28aD-VeGFP expression vector ( figure 1 ). As a control, the two ends of the eGFP gene were added to the BamHI (5′) and SacI (3′) sites to form the p28aD-eGFP expression vector ( figure 1 ). The two plasmids were transformed into Escherichia coli BL21(DE3) star strain to obtain BL28-VeGFP and BL28-eGFP strains, respectively.
Embodiment 3
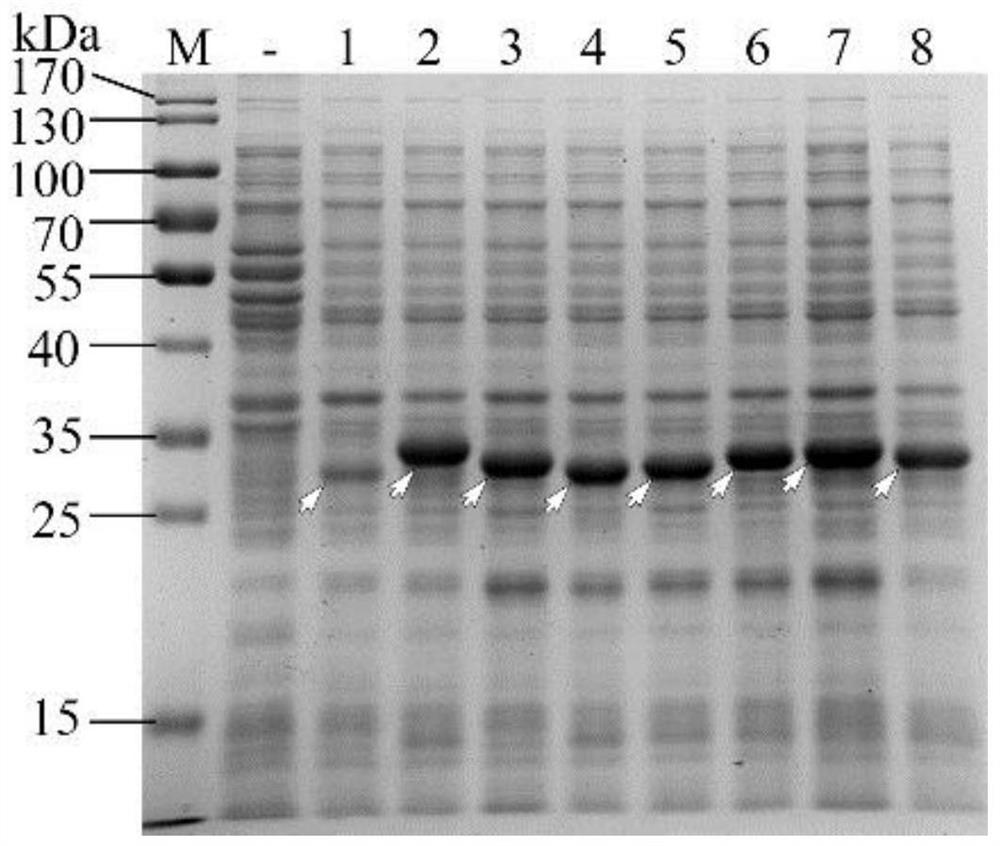
[0039] Embodiment 3: Expression and comparison of VeGFP protein and eGFP protein
[0040] The BL28-VeGFP and BL28-eGFP strains were respectively streaked in solid LB medium (Luria-Bertani medium) containing 50 μg / mL kanamycin, and cultured overnight at 37°C. Select 6 single clones from each strain, pick them up and insert them into the liquid LB medium containing 50 μg / mL kanamycin, shake culture (37° C., 200 rpm) to OD600=0.8. IPTG (isopropylβ-D-1-thiogalactopyranoside) was added to the medium at a final concentration of 1 mmol / L. Under the condition of 16°C, the expression was induced for 16 hours. After induction of expression, pipette 500 μL of each tube of bacterial liquid into a new 1.5 mL centrifuge tube, centrifuge at 12,000 rpm for 5 min, and collect bacterial cells. With 200μL PBS buffer (137mM NaCl, 2.7mM KCl, 10mMNa 2 HPO 4 , and 2mM KH 2 PO 4 , pH 7.4) to resuspend the cells, and add 50 μL of 5× protein loading buffer (250 mM Tris (pH 6.8), 10% (w / v) SDS, 0....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com