Nucleic acid aptamer capable of specifically recognizing florfenicol and florfenicol amine and application of nucleic acid aptamer
A technology of nucleic acid aptamer and florfenicol amine, applied in color/spectral characteristic measurement, biochemical equipment and methods, material inspection products, etc., can solve the problem of difficult to repeat production, antibody preparation requires experimental animals, processing process Complicated problems, to achieve the effect of wide application range, low cost and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
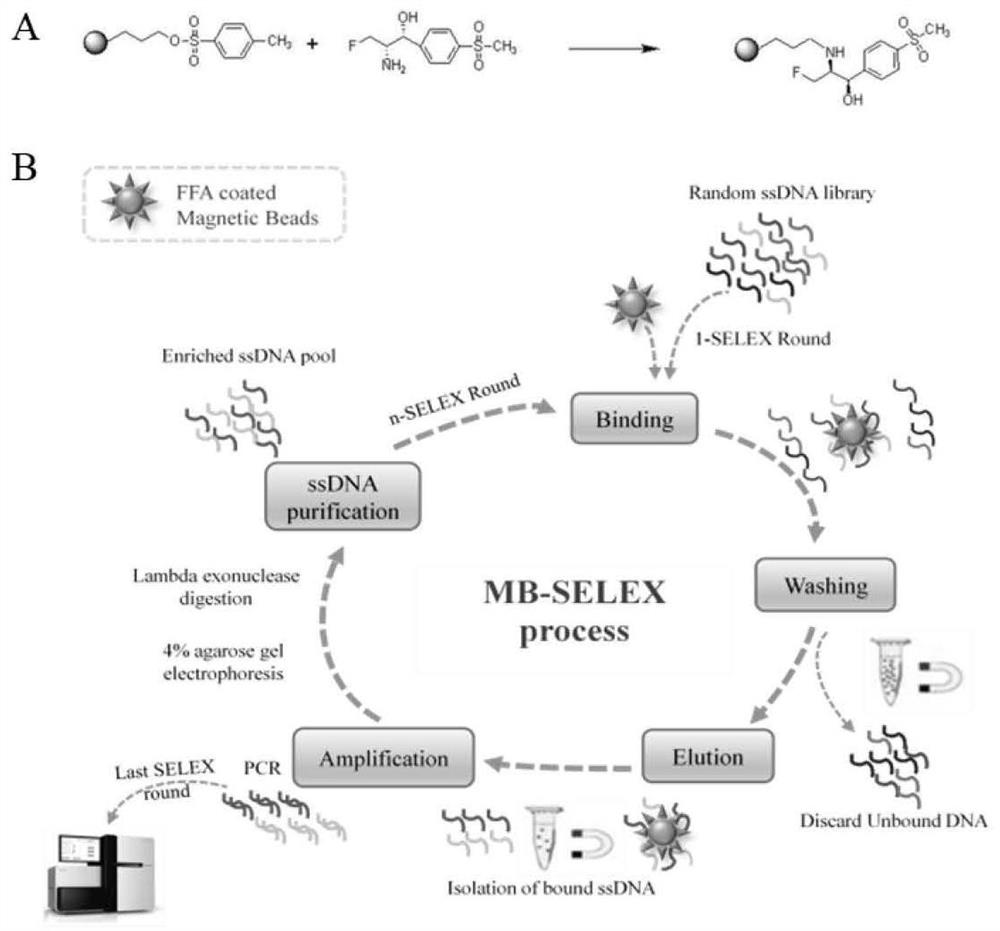
[0055] Example 1. Screening of nucleic acid aptamers that can specifically recognize florfenicol amide
[0056] 1. Synthesize the random ssDNA library and primers shown in the following sequence
[0057] 5'-GCTGTGTGACTCCTGCAA-N 43 -GCAGCTGTATCTTGTCTCC-3'
[0058] Upstream primer: 5'-GCTGTGTGACTCCTGCAA-3'
[0059] Downstream primer: 5'-PHO-GGAGACAAGATACAGCTGC-3'
[0060] Among them, "N 43 "Indicates a sequence formed by connecting 43 arbitrary nucleotide bases. The library and primers were synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.
[0061] The library and primers were stored at -20°C in stock solutions with a concentration of 100 μM prepared from TE buffer (PH=8.0: 10 mmol / L Tris-HCl, 1 mmol / LEDTA) respectively.
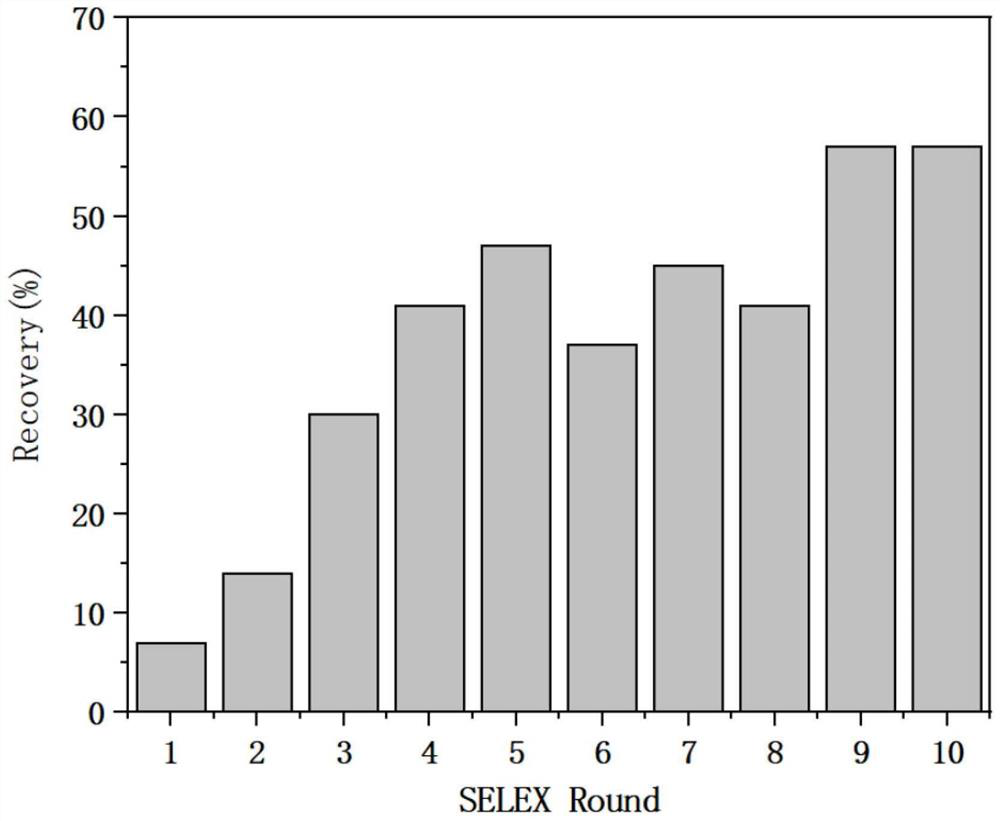
[0062] 2. MB-SELEX screening specific nucleic acid aptamers
[0063] figure 1 It is the covalent coupling (A) and MB-SELEX screening flow chart (B) of tosyl magnetic beads and florfenicol amine provided by the embodiments of the present disclosu...
Embodiment 2
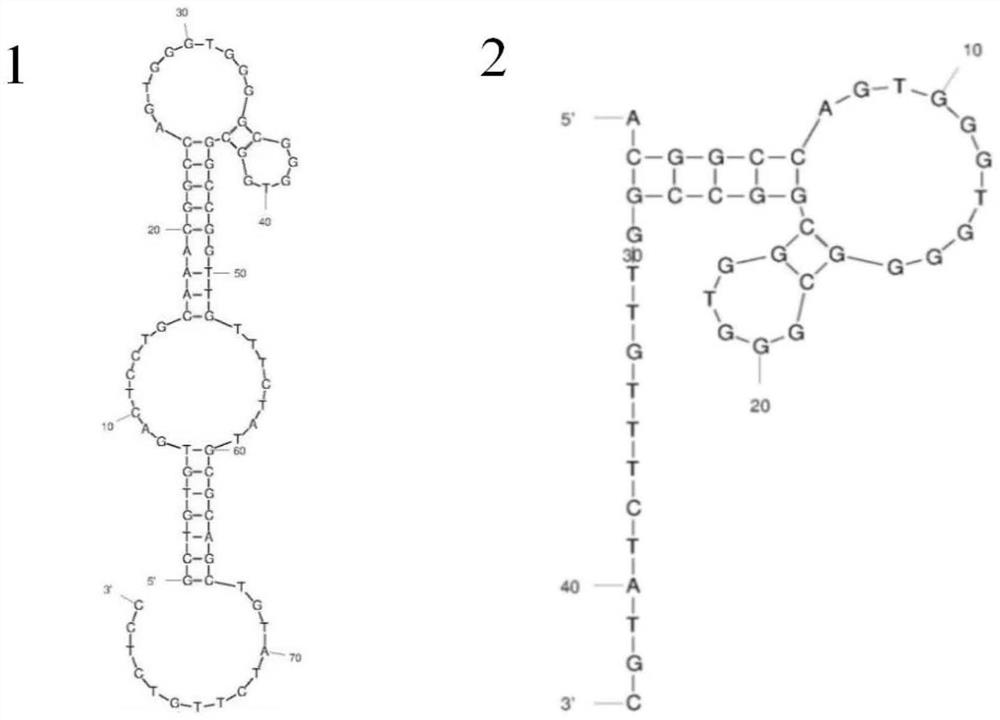
[0097] Embodiment 2, using the short-chain nucleic acid aptamer SEQ ID No.2 obtained after truncation to establish a method for detecting Florfenicol
[0098] Colloidal gold with a particle size of about 20nm was prepared by sodium citrate reduction method. 40μl 1μM nucleic acid aptamer was reacted with 50μl colloidal gold in the dark for 30min, and then a series of concentrations (0.00128ng / mL, 0.032ng / mL, 0.032ng / mL, 0.8ng / mL, 4ng / mL, 100ng / mL, 500ng / mL) of florfenicol, react in the dark at 37°C for 1h, and measure the absorbance value (A650 / A520) by a microplate reader. Take A650 / A520 as the ordinate, and the florfenicol concentration as the abscissa, so as to realize the sensitive detection of florfenicol. The results are as follows: Figure 7 shown. The method has a good linear range in the Florfenicol concentration of 0.00128~500ng / mL, and the minimum detection limit that can detect Florfenicol is 0.00128ng / mL, and the linear regression equation is y=0.0098ln(x)+0.7197...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com