Primer pair for identifying carrot petal cytoplasmic male sterility and application thereof
A technology for male sterility and male sterility, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problems of complex genetic background of male sterility in carrots, etc. The effect of reliable technical means
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
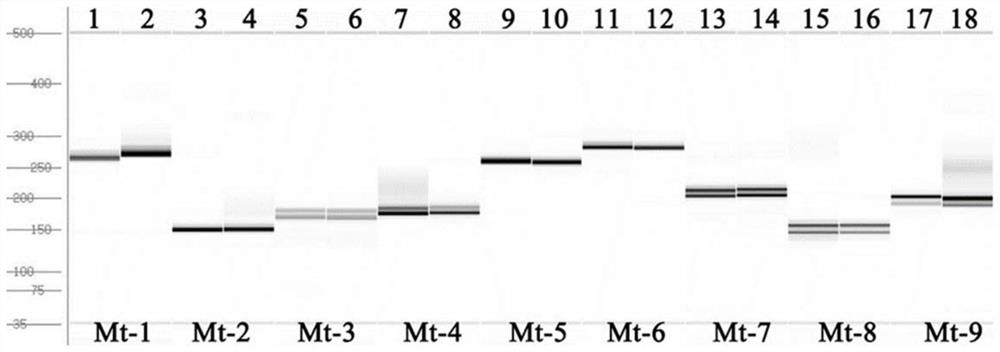
[0052] A group of inflorescence RNAs from carrot petal cytoplasmic male sterile lines and their maintainers (HYs698 and HYd698) of the Hongyue strain bred by the Vegetable Research Institute of Tianjin Academy of Agricultural Sciences were extracted, a cDNA library was established, and the transcriptome was sequenced. , compared with the carrot reference mitochondrial genome, the URL of the reference mitochondrial genome is as follows: http: / / ftp.ensemblgenomes.org / pub / plants / release-51 / fasta / daucus_carota / dna / ; Screen out 32 positions with base number differences (Indel≥4bp) between fertile and sterile, as shown in Table 1 for schematic examples.
[0053] Table 1 Nine differential positions screened by transcriptome analysis
[0054]
[0055] On the basis of including this difference site, the sequence was extended up and down by about 100 bp each, and 9 pairs of primers were designed in order to amplify the bands, corresponding to the difference positions 1-9 respective...
Embodiment 2
[0061] (1) Sample group 1: WMA-477 and WMB-477 are derived from Japanese Kuroda-type carrot petal-type cytoplasmic male sterile lines and their maintainers; LSsA-128 and LSsB-128 are derived from American cut-type carrots The petalized cytoplasmic male sterile line and its maintainer line; SKs35-027 and SKd35-027 are derived from the carrot petalized cytoplasmic male sterile line and its maintainer line with the combination of Nantes and Kuroda; HYs698 and HYd698 are derived from Carrot petal-type cytoplasmic male sterile line and its maintainer line of Hongyue strain bred by Vegetable Research Institute of Tianjin Academy of Agricultural Sciences.
[0062] (2) Reagents: 2×Taq Master Mix (for PAGE); specific amplification primers synthesized by Jinweizhi.
[0063] The forward primer sequence is SEQ ID NO.1: 5'-TTGTGCGGTTAAGGTTGTTGT-3'
[0064] The reverse primer sequence is SEQ ID NO.2: 5'-CCTGTTCTTGTGGATCAATGGA-3'.
[0065] (3) Polymorphism detection: extract the genomic DN...
Embodiment 3
[0075] (1) Sample group 2: 8 commercial varieties available on the market (Mendel No. 3, SK316, Chunao 709, Nobel No. 2, Rose Red, Laxian, Chunao 705, Hongbao Six-inch Ginseng).
[0076] (2) Reagents: 2×Taq Master Mix (for PAGE); specific amplification primers synthesized by Jinweizhi.
[0077] The forward primer sequence is SEQ ID NO.1: 5'-TTGTGCGGTTAAGGTTGTTGT-3'
[0078] The reverse primer sequence is SEQ ID NO.2: 5'-CCTGTTCTTGTGGATCAATGGA-3'.
[0079] (3) Polymorphism detection: extract the genomic DNA of young carrot leaves to be tested, and use it as a template to perform PCR amplification using the specific primer pair provided by the present invention.
[0080] (4) PCR reaction system: a total volume of 10 μL, including 5 μL of 2×Taq Master Mix (for PAGE), 0.5 μL of upstream and downstream primers (10 μmol / L), 1 μL of DNA template (25ng / μL), supplemented with sterile water to 10 μL.
[0081] (5) PCR amplification program: pre-denaturation at 95°C for 5 min, 1 cycle;...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com