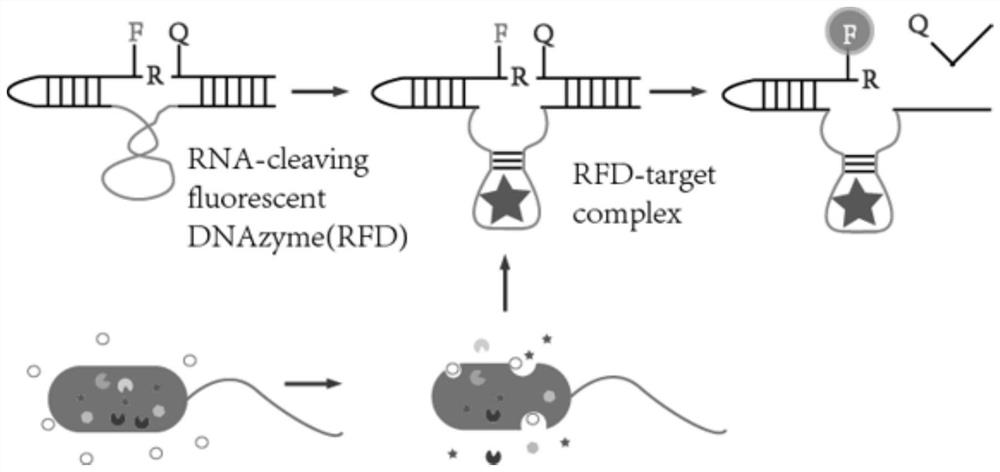
Application of deoxyribozyme probe in escherichia coli drug-resistant phenotype high-throughput sensing
A DNAzyme probe, Escherichia coli technology, applied in the field of analysis and detection, can solve problems such as inability to achieve phenotype analysis, and achieve the effects of preventing the formation of multidrug-resistant bacteria, accurate sensing results, and preventing the abuse of antibiotics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 Rapid detection of tetracycline-resistant Escherichia coli using RFD-EC recognition molecules
[0040] Rapid detection of tetracycline-resistant Escherichia coli using RFD-EC recognition molecules, the principle is as follows figure 2 As shown, the specific operation is as follows:
[0041] a) Cultivate engineering bacteria of tetracycline-resistant Escherichia coli, add 10 μL of tetracycline-resistant bacteria to 5 mL of SOB medium, and carry out overnight culture, the temperature of the shaking table is 37 ° C, and the shaking speed is 150 rpm;
[0042] b) After culturing overnight, use a microplate reader to measure the bacterial concentration of Escherichia coli, and when OD600=0.5, the bacteria are taken out from the shaker, and the bacterial concentration is about 10 8 CFU / mL;
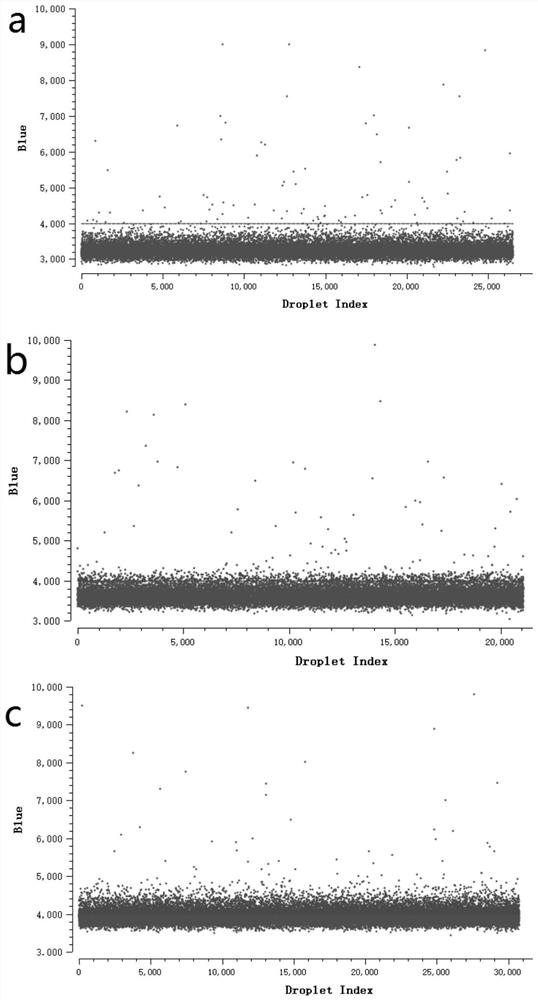
[0043] c) Dilute the tetracycline-resistant Escherichia coli with sterile water so that the bacterial concentration is 10 5 CFU / mL; by calculation in a 10.5 μL bacterial samp...
Embodiment 2
[0049] Example 2 Rapid determination of kanamycin-resistant Escherichia coli using RFD-EC recognition molecules, and bacterial counting
[0050] Using RFD-EC recognition molecules to quickly detect kanamycin-resistant Escherichia coli, the specific operation is as follows:
[0051] a) Cultivate engineering bacteria of kanamycin-resistant Escherichia coli, add 10 μL of kanamycin-resistant bacteria to 5 mL of SOB medium, and carry out overnight culture, the temperature of the shaking table is 37 ° C, and the shaking speed is 150 rpm;
[0052] b) After culturing overnight, use a microplate reader to measure the bacterial concentration of Escherichia coli, and when OD600=0.5, the bacteria are taken out from the shaker, and the bacterial concentration is about 10 8 CFU / mL;
[0053] c) Dilute kanamycin-resistant Escherichia coli with sterile water so that the bacterial concentration is 10 4 ~10 6 CFU / mL, by calculation in a 10.5 μL bacterial sample, the number of bacteria is 100,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com