Rare genetic disease mutant gene and application thereof
A technology for mutated genes and genetic diseases, applied in the field of mutated genes for rare genetic diseases and its applications, can solve the problems of GL-3 degradation blockage, α-GalA enzyme function loss, ischemia, etc., and achieve the effect of reducing the birth of children
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Example 1 - Mutated Genes for Rare Inherited Diseases
[0018] Rare genetic disease mutated genes, the specific mutations are shown in Table 1 below:
[0019] Table 1 Specific detection results of mutated genes in rare genetic diseases
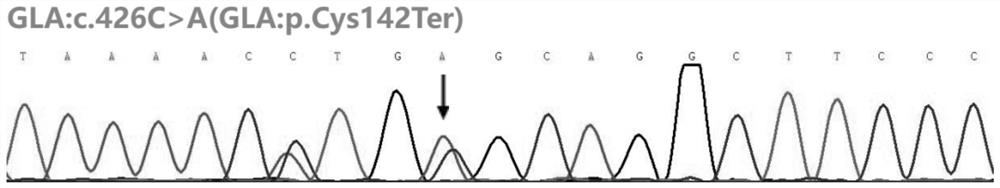
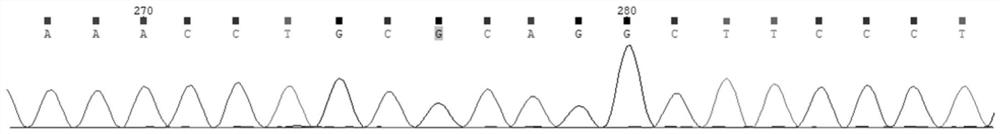
[0020] Gene genomic location transcript number base change amino acid changes Reference Genome Version exon number GLA chrX:100656741 NM_000169 c.426C>A p.Cys142Ter GRCh37 / hg19 Exon3
[0021] (1) At genomic position chrX:100656692-chrX:100656791, the sequence of the wild-type GLA gene is:
[0022] is the base of the wild-type GLA gene at chrX:100656741 in the genome.
[0023] At the corresponding genome position, the sequence of the rare genetic disease mutation gene is:
[0024] is the base of the mutant gene GLA at chrX:100656741 in the genome.
[0025] (2) When the transcript number is NM_000169, the reference sequence of the DNA encoding the wild-type GLA gene is:
[0026] ...
Embodiment 2
[0032] Example 2-Detection kit for rare genetic disease mutation gene
[0033] Detection kits for mutation genes of rare genetic diseases, including Taq DNA polymerase, PCR buffer and primers, etc. The specific primers are as follows:
[0034] Upstream primer (GLA-E16F, SEQ ID NO: 1): 5'CTGCTACCTCACGATTGTGCT 3';
[0035] Downstream primer (GLA-E16R, SEQ ID NO:2): 5'ACCTGGACTCCCCTAA 3';
[0036] Length: 532bp.
[0037] The specific steps of using this kit to screen the mutated pathogenic gene GLA are as follows: extract the DNA of the test subject, and then use the designed primer combination (SEQ ID NO: 1 and SEQ ID NO: 2) to amplify the GLA gene to obtain For the PCR product, use 1.5% agarose gel electrophoresis to detect the PCR product, select 1000bp Marker as a reference, check and verify that the amplified product is the expected size, and finally sequence the PCR product. Obtained reference sequences from the NCBI (https: / / www.ncbi.nlm.nih.gov / ) database and compared...
Embodiment 3
[0038] Embodiment 3-family verification experiment
[0039] In this example, the method of family linkage analysis is used to verify the pathogenicity of mutant genes of rare genetic diseases.
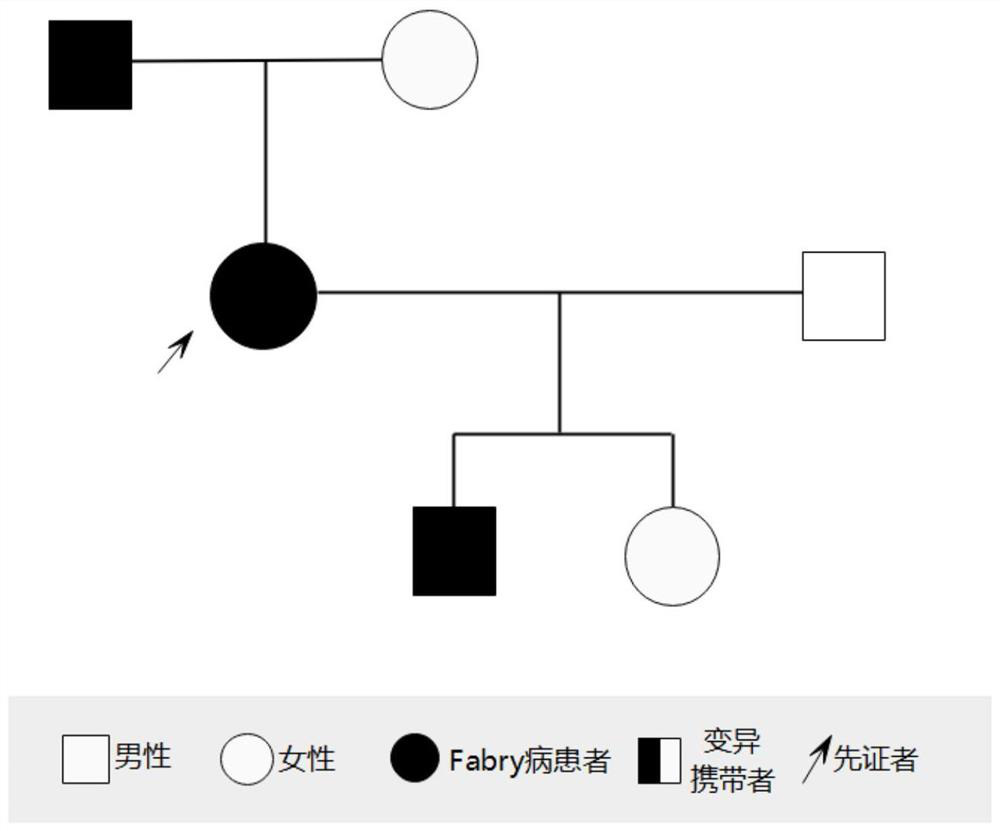
[0040] Specifically, three generations of members of a family with familial Fabry disease were selected, and the proband (female, 57 years old) in this family was clinically diagnosed with Fabry disease.
[0041] On the premise that the proband and his family members voluntarily sign the informed consent, 5-10mL whole blood samples will be sent, and a medical record database will be established to record the proband's condition and family status in detail. This study has been approved by the institutional ethics committee.
[0042] Description of the clinical profile of the proband:
[0043] Table 3 Clinical profile of the proband
[0044]
[0045] The GLA gene of the proband and his family members was tested using the in vitro detection kit provided in Example 2, and the results...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com