Fluorescence in-situ hybridization probe for identifying group B chromosomes of triticum aestivum as well as design method and application of fluorescence in-situ hybridization probe
A fluorescence in situ hybridization and chromosome technology, applied in the field of molecular cytogenetics, can solve the problems of insufficient signal, clear, wrong identification, inaccuracy, etc., to achieve good application prospects, save time, and improve experimental efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] 1. Development of oligonucleotide probes
[0069] (1) Download the wheat genome data from the NCBI database, and use MISA software to analyze the SSR repeat sequence of the above data, and obtain different types of wheat SSR repeat sequences.
[0070] (2) Select the SSR with a large number of repetitions as the candidate sequence. The obtained candidate SSR sequences were compared and verified by the online visual comparison website B2DSC (http: / / mcgb.uestc.edu.cn / b2dsc), and the SSR sequences with the appropriate size (20-60bp) of wheat fragments were selected.
[0071] (3) Obtain the SSR sequence shown in the sequence listing.
[0072] 2. Probe Preparation
[0073] The SSR sequence designed in 1 was sent to Thermo Fisher Scientific (Shanghai) Co., Ltd. to synthesize an oligonucleotide probe, and its 5' end was fluorescently labeled with 6-FAM.
Embodiment 2
[0075] 1. Wheat root tip chromosome preparation
[0076] (1) Put the common wheat seeds into a petri dish and soak them in distilled water for 6 hours, put them on filter paper to germinate, and wash them twice a day with distilled water.
[0077] (2) When the root grows to 1.5-2cm, pour off the excess water in the culture dish, cut off the root tip and put it into a 0.5mL centrifuge tube that has been punched in the lid and sprayed with distilled water to wet the tube wall, and then put the centrifuge tube into filled with N 2 O gas in a closed metal tank for 2h.
[0078] (3) Take out the centrifuge tube, add 90% acetic acid to fix it for 5 minutes, and rinse it with distilled water 3 times.
[0079] (4) Cut the roots cleaned with distilled water and take the root tip meristematic zone, put them into 1% pectinase and 2% cellulase solution, and put them in a water bath at 37°C for 50 minutes for enzymatic hydrolysis.
[0080] (5) The enzymatically hydrolyzed root tip was wa...
Embodiment 3
[0084] 1. FISH test
[0085] (1) Prepare a hybridization solution system, each slide contains probe 0.8 μL, ssDNA 4 μL, 2×SSC+1×TE 5.2 μL.
[0086] (2) Put the slides with clear chromosomes into the ultraviolet cross-linking instrument for 2-3 times of cross-linking.
[0087] (3) 10 μL of hybridization solution was dropped on each slide, covered with a cover glass and placed in a humid aluminum box for hybridization at 42°C for 2 hours.
[0088] (4) Rinse the coverslips with 2×SSC and dry them, add 20 μL DAPI dropwise to each slide, and cover the coverslips.
[0089] Further, the DAPI solution is obtained by diluting the DAPI stock solution (VECTOR, USA) 3 times with 1×PBS.
[0090] (5) Slides were placed under an Olympus BX60 fluorescence microscope and photographed with a Cellsens Standard camera system.
[0091] 2. Result identification
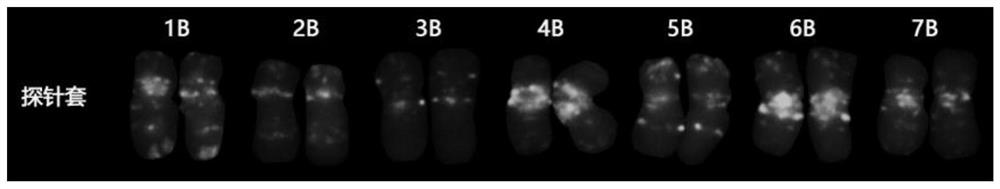
[0092] Probe first (ATGTTG) 4 Do the first FISH, and observe the photographs, such as figure 1 shown; following signal elution on t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com