Method for rapidly identifying gene for regulating and controlling oil content of rape based on genomics approach and application thereof
A genomics and oil content technology, applied in chemical instruments and methods, botanical equipment and methods, biochemical equipment and methods, etc. Good application prospects, the effect of shortened time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1 Rapid Identification of Genes Regulating Rapeseed Oil Content Based on Genomics Approach
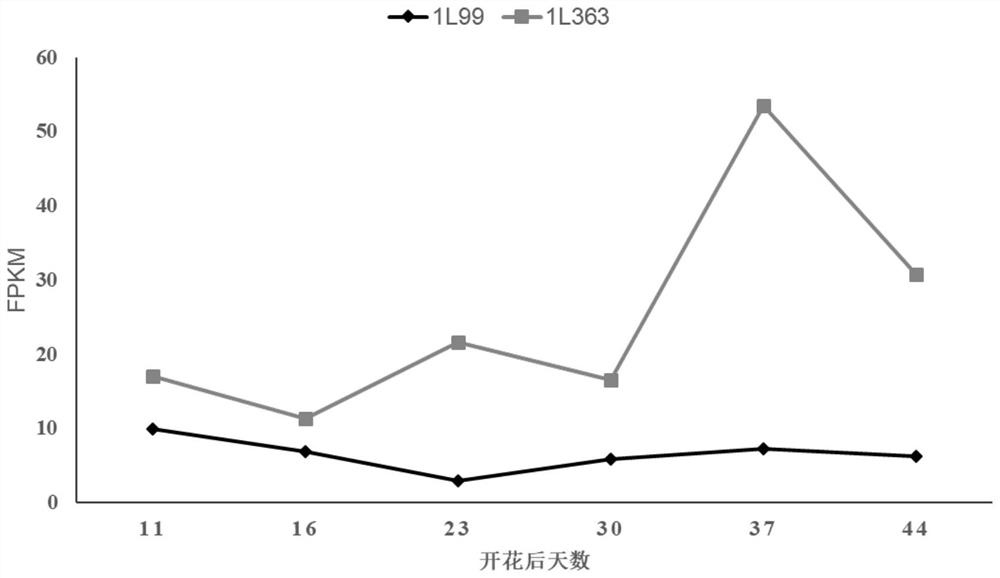
[0032] 1. RNA sample preparation of high oil content inbred line 1L99 and low oil content inbred line 1L363
[0033] Brassica napus high oil content inbred line 1L99 and low oil content inbred line 1L363 were planted in the experimental field of Huazhong Agricultural University during the sowing season. The siliques at different developmental stages were collected at 11 days, 16 days, 23 days, 30 days, 37 days and 44 days after flowering, and three biological replicates were taken for samples at each stage. According to the instructions of the RNA extraction kit (TransZol Plant, BioTeke, China), the sample RNA was extracted, and the RNA was treated with RNase-free DNase-I (Thermo Scientific, USA) to remove gDNA contamination, and a nucleic acid / protein analyzer was used 730 (Beckman ) for quantification.
[0034] 2. Library construction, sequencing and analysis
[...
Embodiment 2
[0038] Example 2 Verification of the gene function of BnECH.A5 by CRISPR / Cas9-mediated gene knockout technology
[0039] 1. Screen target sequences and design primers
[0040] The nucleotide sequence (such as SEQ ID NO. 17) of the Brassica napus BnECH.A5 gene was obtained from the reference genome Darmor-bzh. Two targets, T1 and T2, were designed and screened (Table 1), and primers were designed for the target sequences (Table 2).
[0041] Table 1 Target sequence
[0042]
[0043] Table 2 target primer sequence
[0044]
[0045]
[0046] 2. Construction of the two-target gene editing vector pBnECH vector
[0047] (1) Target linker preparation: Dissolve the linker primers in Table 2 with ddH2O into a 10 μM stock solution. Add 1 μl of the left and right primers to 8 μl ddH2O, mix and dilute to 1 μM, and vortex to mix. Then put it into a water bath at 90°C, treat it for 30", and let it cool down to room temperature naturally.
[0048] (2) sgRNA expression cassette ...
Embodiment 3
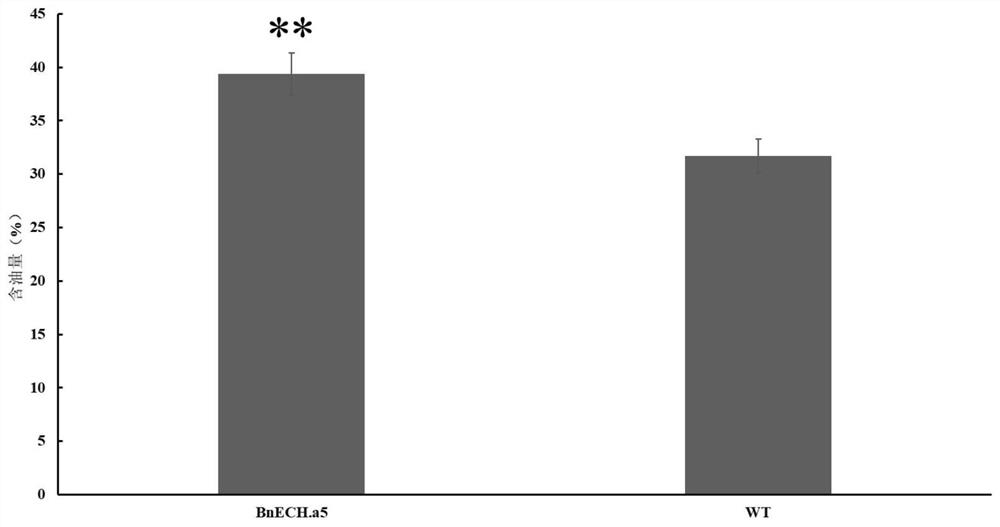
[0134] Example 3 Rapeseed BnECH.A5 mutation can be used to increase seed oil content
[0135] After the mutant plants were obtained in Example 2, the homozygous mutant plants were obtained by selfing.
[0136] The oil content of mutant seeds was determined as follows.
[0137] Sample preparation:
[0138] 1) The mutant and wild-type seeds were dried in a constant temperature incubator at 37°C for 48 hours to ensure the consistency of seed moisture content. Each individual plant took out 50 mg seeds of uniform size and weighed and recorded the weight for subsequent use;
[0139] 2) Crush the weighed rape seeds with a mortar, place them in a clean 10ml glass test tube, and spin the seeds to the bottom of the tube by centrifugation;
[0140] 3) Add 1.5ml 2.5% sulfuric acid chromatographically pure methanol solution (0.01% BHT), 200μl standard sample (heptadecanoic acid triglyceride, 2mg / ml), 400μl chromatographically pure toluene to the above glass tube, and tighten the screw ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com