Tobramycin detection system and method based on CRISPR-Cas12a
A technology of tobramycin and detection system, applied in measurement devices, instruments, fluorescence/phosphorescence, etc., can solve the problems of high false positive rate, long operation time, lack of ultraviolet chromophore, etc. The effect of visualization and quantitative detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035]A CRISPR-Cas12a-based tobramycin detection system, including: aptamer probe AP, CrRNA, AsCas12a, KF polymerase, and reporter probes modified with fluorescent groups and quenching groups at both ends;
[0036] The sequence of the aptamer probe AP is: 5'-ATC ATT TGG AGG AAC TGG AGT CAC AAG CTGAGG ATG TGA CTC CAG GCA CTT AGT CAC A-3' (SEQ ID NO: 1);
[0037] The sequence of the CrRNA is: 5'-UAA UUU CUA CUC UUG UAG AUG CUU GUG ACU CCA GUUCCU C-3' (SEQ ID NO: 2);
[0038] The sequence of the reporter probe is: 5'-FAM-TTATT-BHQ1-3'.
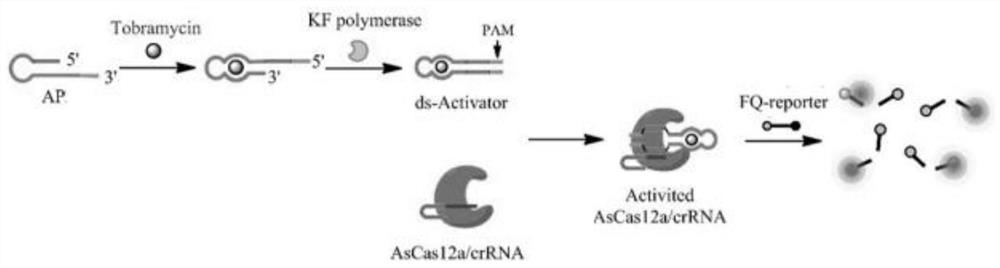
[0039] The detection principle diagram of the tobramycin detection system of the present invention is shown in figure 1 . The designed aptamer probe AP of the present invention comprises two functional areas: one is the aptamer sequence of tobramycin, and the other is a signal transduction sequence used for KF polymerase binding and extension for signal amplification, These two regions are designed to partially hybridize to form a stem-loop st...
Embodiment 2
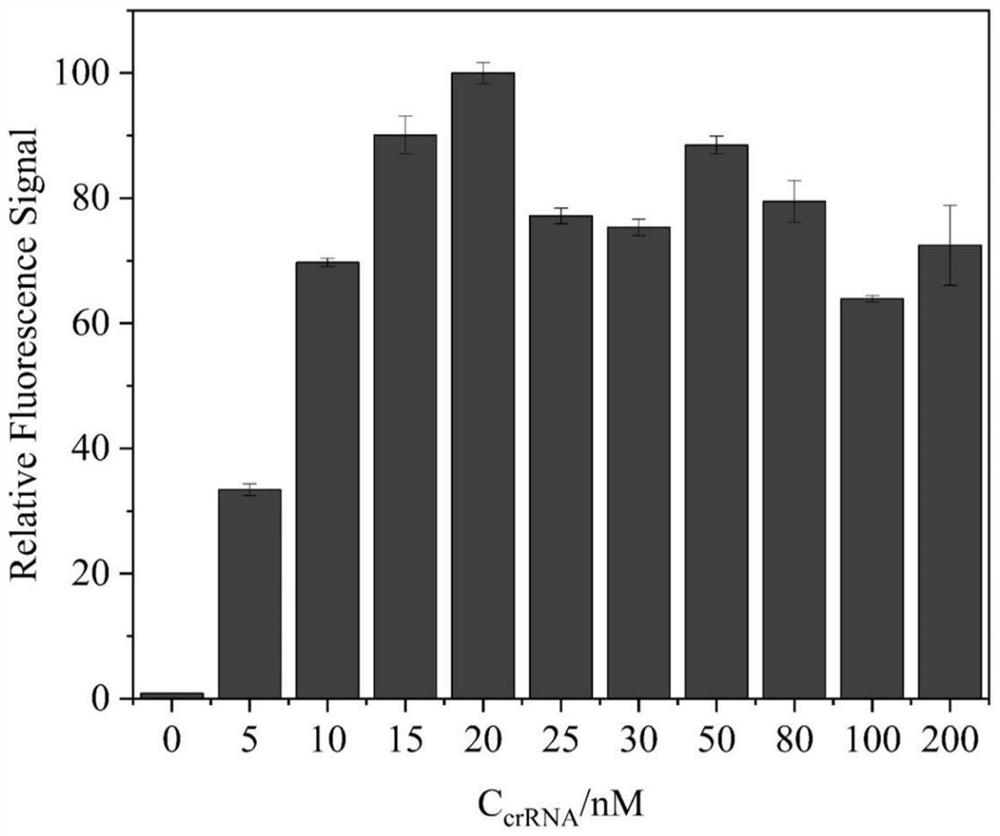
[0049] The present invention uses the CRISPR-Cas12a system as the signal output element, and the concentration of crRNA directly leads to the sensitivity of the CRISPR-Cas12a system to recognize the trigger chain. In order to explore the optimal crRNA concentration of the CRISPR-Cas12a system, different concentrations of crRNA (0nM, 5nM, 10nM, 15nM, 20nM, 25nM, 30nM, 50nM, 80nM, 100nM, 200nM), constant double-strand trigger strand (ds- Activator) at a concentration of 1 nM. The result is as figure 2 As shown, the fluorescence intensity reaches the highest when the crRNA concentration is 20nM, and the fluorescence intensity will decrease when it is greater than 20nM, so the crRNA concentration of 20nM is used for analysis in the present invention to ensure higher detection efficiency.
Embodiment 3
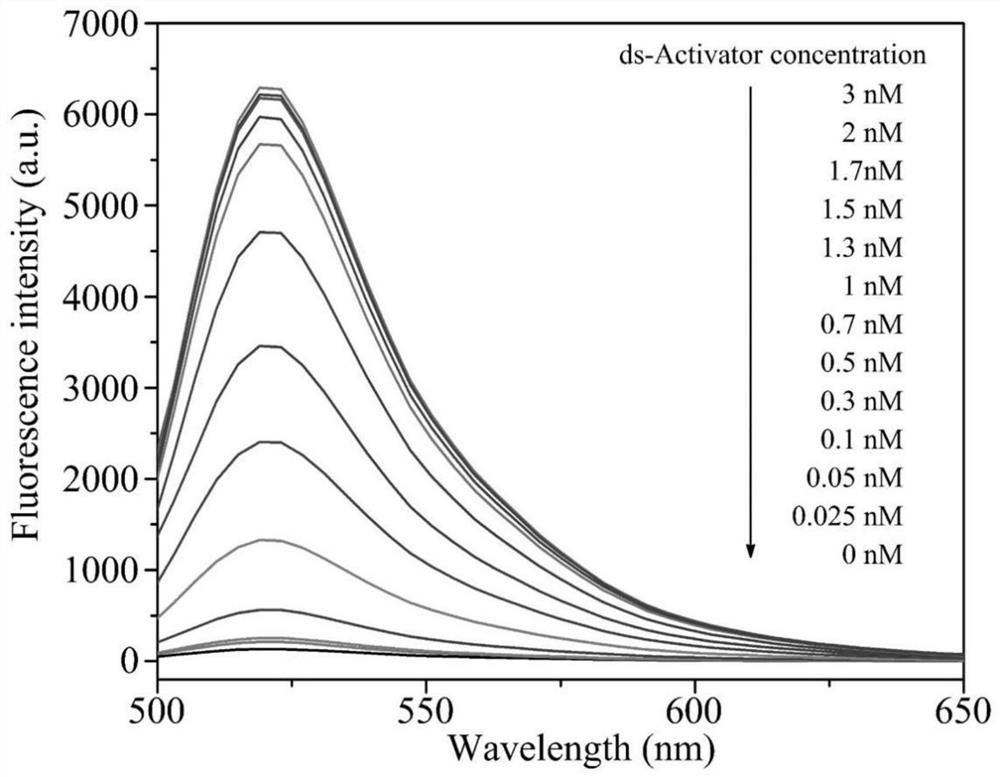
[0051] The response of the CRISPR-Cas12a system to different concentrations of the trigger chain: In order to verify the characteristics of the CRISPR-Cas12a system, different concentrations of the trigger chain (ds-Activator) were designed to explore the concentration response range and the minimum concentration of the CRISPR-Cas12a system to the trigger chain. The concentration of the response. Such as image 3 The fluorescence intensity curves generated by the CRISPR-Cas12a system under different concentrations of the trigger chain. It can be seen that when the excitation wavelength is 480nm, there is an emission peak at 520nm. According to the fluorescence intensity at 520nm, the correlation between the fluorescence intensity measured within the trigger chain concentration range and the trigger chain concentration can be drawn. The result is as follows Figure 4 shown. Figure 4 The inset of the graph shows a linear relationship between the fluorescence intensity and the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com