CRISPR/Cpf1 gene editing system, as well as construction method of CRISPR/Cpf1 gene editing system and application of CRISPR/Cpf1 gene editing system in gibberella
A gene editing and construction method technology, applied in the field of genetic engineering, can solve the problems of low efficiency of multi-gene editing, speed up the production of bacterial strains, and low efficiency of integration technology, and achieve the effect of simple system, easy repair, and favorable plasmid transformation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0090] 1. Amplification of genetic elements and preparation of target plasmids
[0091] 1. Preparation of target gene
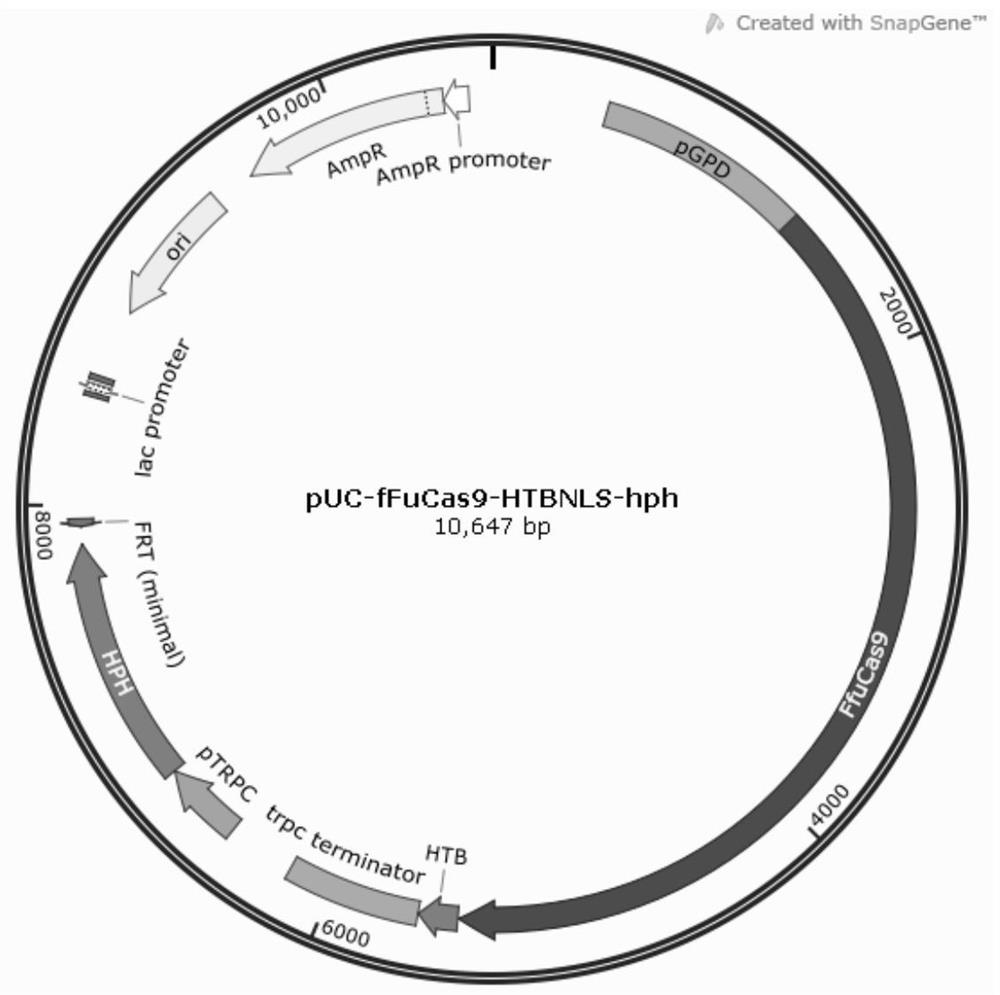
[0092] The plasmid pUC-fFuCas9-HTBNLS-hph (SEQ ID NO.25) ( figure 1) as a template, design primers HTBnls-R (SEQ ID NO.7) and HTBnls-F (SEQ ID NO.8), amplify the plasmid, and obtain a backbone linear fragment that does not contain the fFuCas9 gene, which is used as a subsequent recombinant plasmid skeleton.
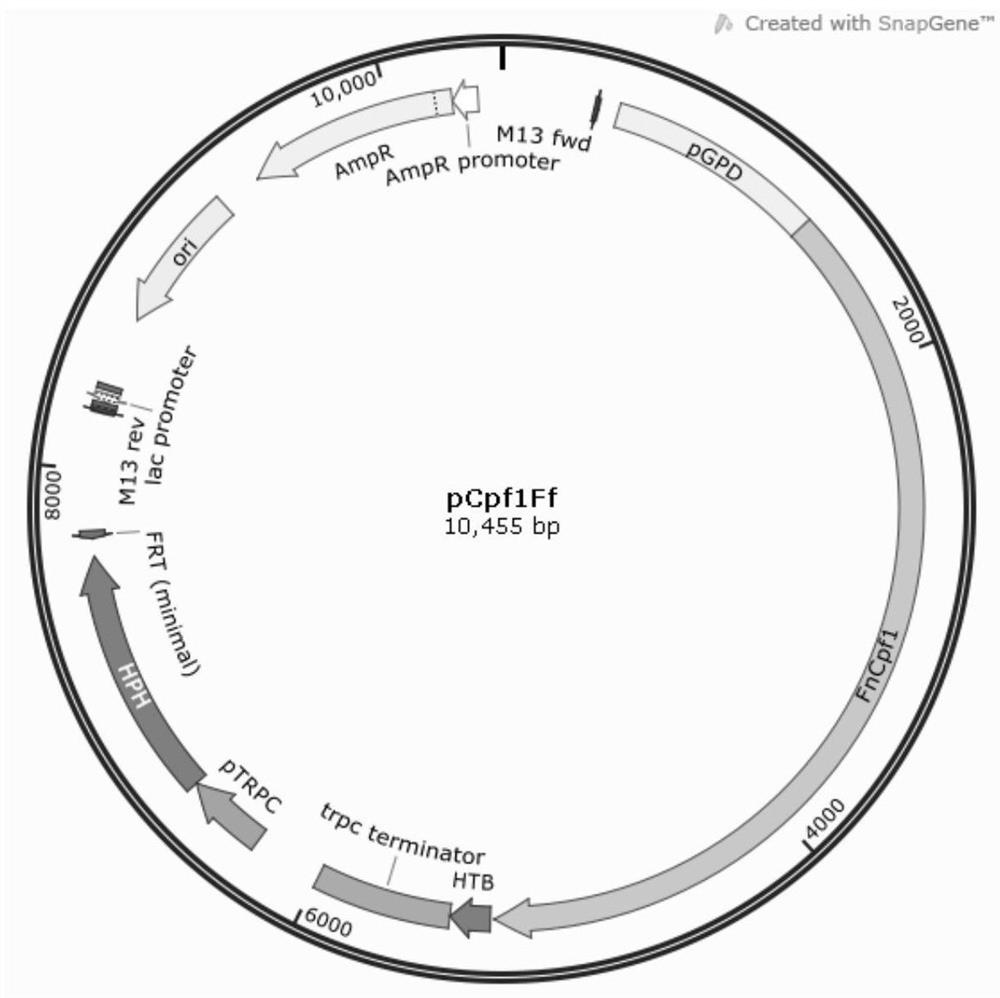
[0093] According to the Fncpf1 sequence provided on NCB1, Qingke Biotechnology Co., Ltd. was commissioned to synthesize Fncpf1. The Fncpf1 nucleotide sequence is shown in SEQ ID NO.2. The Fncpf1 gene and the above-mentioned backbone linear fragment were cloned in one step through the ClonExpressMultiS One Step Cloning Kit. The plasmid pCpf1Ff was constructed, which contained promoter pGPD (SEQ ID NO.1)-Fncpf1-terminator trpc-promoter pTRPC (SEQ ID NO.5)-hygromycin selection marker hph (SEQ ID NO.6) expression frame( figure 2 ). The plasmid pCpf1Ff...
Embodiment 2
[0153] Preparation of Gibberella Competent
[0154] (1) A single colony of Gibberella was inoculated into 100 mL of regeneration medium, cultured at 30° C. and rotated at 200 RPM, and cultivated for 48 hours. (2) Inoculate 10 mL of the bacterial solution into a new 100 mL regeneration medium and expand the culture for 12 hours. (3) The culture solution was filtered with sterile lens tissue to collect the bacteria. (4) Prepare 10mL enzymatic hydrolysis solution: dissolve the mixed enzyme (45mg, 105mg) with a mass ratio of Dryalase (collapse enzyme): Lysing (lysing enzyme) = 3:7 in sodium chloride buffer (0.41g in 10mL) , stir well. After sterilizing with a 0.22μm filter head, add 1g of bacterial cells per 10mL, 30°C, 200RPM, and enzymolysis for 3.5h. (5) The enzymatic solution was filtered through 3 layers of lens tissue to remove mycelium fragments. (6) The protoplast solution was obtained by filtration, centrifuged at 3000 RPM for 10 min, and the protoplast was precipitat...
Embodiment 3
[0156] Transformation of recombinant plasmids
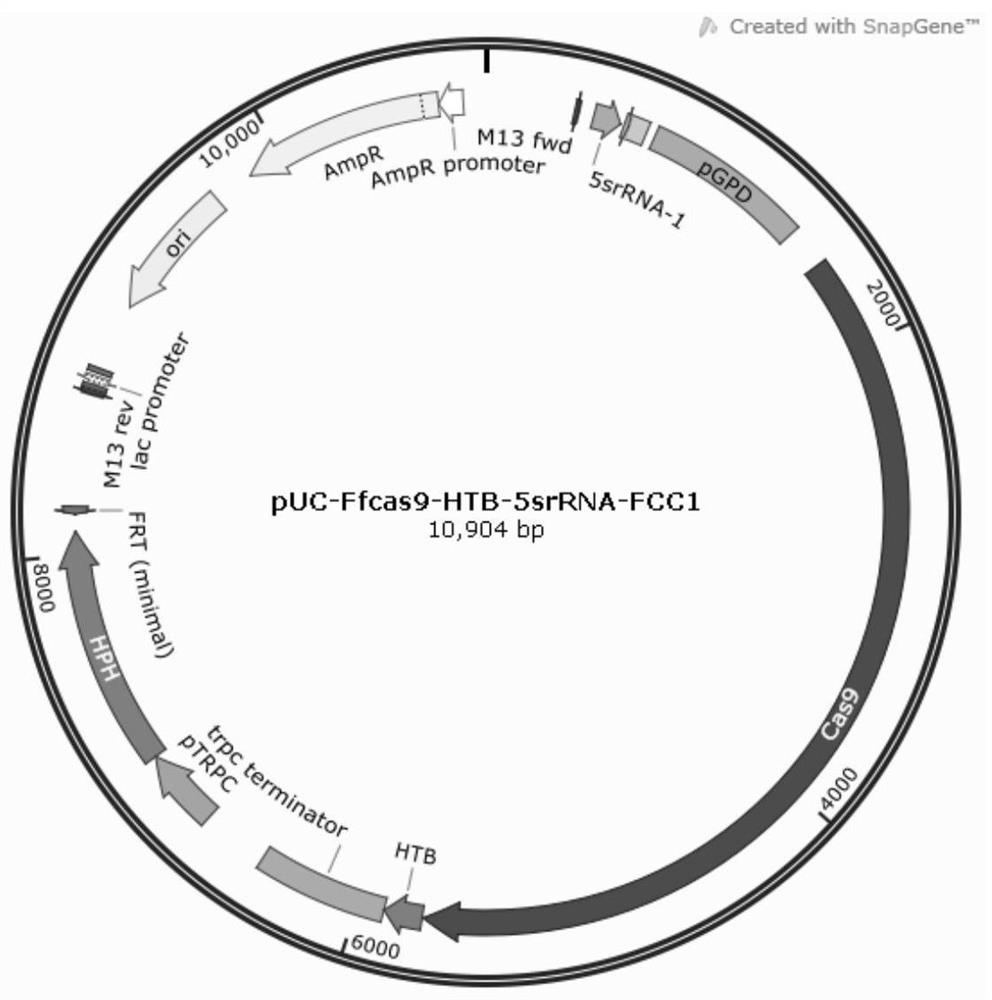
[0157] (1) Take 100 uL of the protoplasts of Example 2, add 50 μL of STC solution, add 10 μg of the recombinant plasmid pCpf1Ff-DR-FCC1 obtained in Example 1, mix and ice-bath for 20 minutes. (2) Add 50 μL of 25% PEG6000 solution (1M sorbitol, 10mM Tris-Cl, 50mM CaCl 2 , pH=7.5) and ice bath for 20 minutes after mixing. (3) Add 1mL PEG6000 solution, mix at room temperature for 5min, then add 2mL STC solution, mix well, take 160μL of MYG solid regeneration plate, incubate for 12h, cover 10mL hygromycin soft agar regeneration plate with a concentration of 100μg / ml and wait Transformants appeared in 5-7 days. The expected purple fungus was obtained on the plate (Gibberella itself is white, which is the phenomenon that FCC1 is knocked out), and the genome sequencing results show that the FCC1 gene has been successfully knocked out.
[0158] By transforming the recombinant plasmid pCpf1Ff-DR-FCC1 into Gibberella competently coating...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com