A prediction method of lactic acid bacteria antimicrobial peptides based on graph neural network
A technology of neural network and prediction method, which is applied in the field of identification of biological antimicrobial peptides, can solve problems such as time-consuming, accuracy improvement, and incapable of high-throughput identification, and achieve the effect of improving identification accuracy index and realizing batch identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0023] The technical solutions of the present invention will be further described below with reference to the accompanying drawings and embodiments.
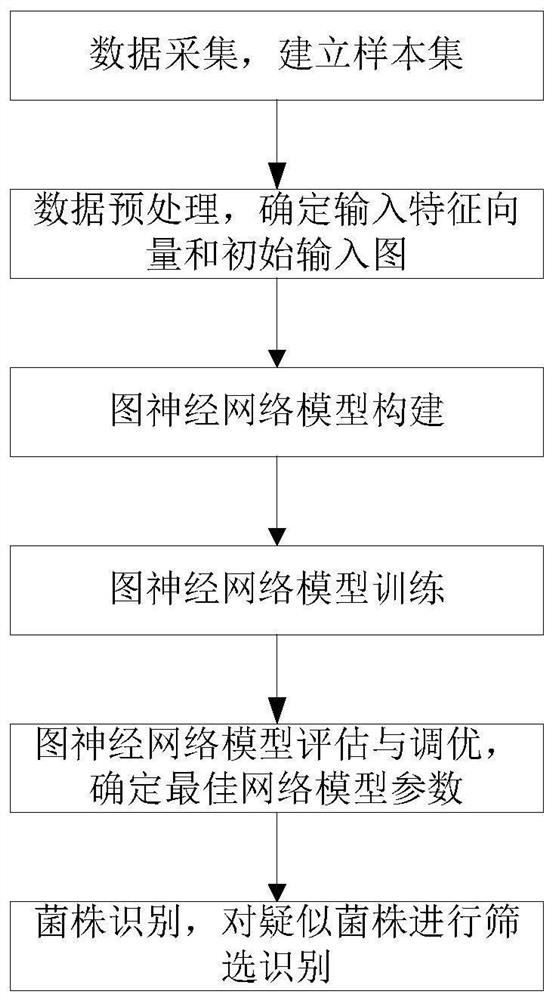
[0024] As shown in the figure, a prediction method of lactic acid bacteria antimicrobial peptides based on graph neural network is mainly divided into four aspects: data collection, model establishment, model tuning, and model prediction.
[0025] Specifically, it can be subdivided into the following steps:
[0026] S1, data collection, establish positive samples and negative samples
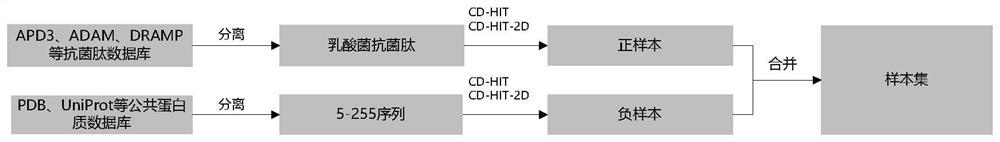
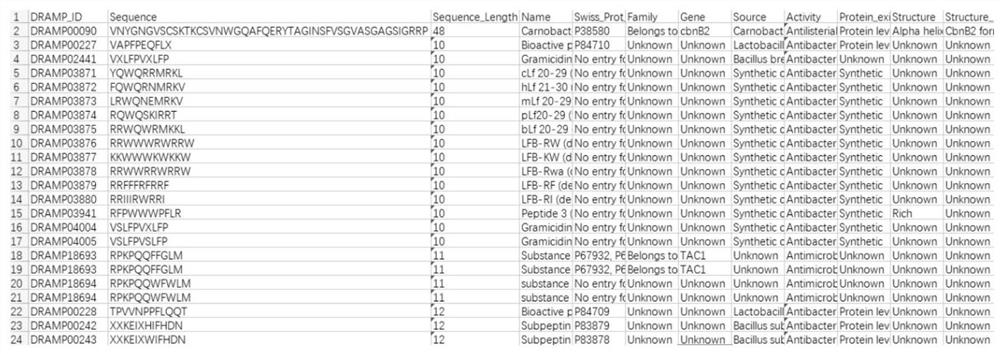
[0027] The positive sample is the collection of lactic acid bacteria antimicrobial peptide sequences isolated in the comprehensive and special antimicrobial peptide database obtained by the investigation, and the negative sample is the collection of protein sequences that meet the length requirement of 5-255. The sample set is established based on the positive and negative samples.
[0028] like figure 2 As shown, Lactobacillus antibacterial pe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com